Introduction
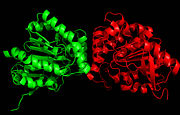
Figure 1. Ag85C homodimer.
Mycobacterium tuberculosis is the bacteria that causes the tuberculosis (TB) disease itself, a leading infectious cause of death world-wide. Thus, it is necessary to develop antimycobacterial drugs. However, this has proven to be difficult as the bacteria can become resistant as a result of misuse of drugs. This gives the bacteria time to mutate which ultimately leads to the drug resistance. To combat this, scientists and researchers have taken on many studies and clinical trials to locate specific drug targets. Obstacles for controlling TB infection include lengthy treatment regimens, drug resistance, lack of a highly efficacious vaccine, and incomplete understanding of the factors that control virulence and disease progression.[1] Of these projects, some deal specifically with the M. tuberculosis Antigen 85 (Ag85) complex. Ag85 complex consists of 3 secreted enzymes (A, B, and C) and plays a key role in pathogenesis and cell wall synthesis of M. tuberculosis. [2]
The cell wall of mycobacterium tuberculosis covalently linked molecules, one of which is mycolic acid, a fatty acid which exists in the membrane as the free glycolipids trehalose monomycolate (TMM) and trehalose dimycolate (TDM). Studies suggest that these three components are necessary to maintain the integrity of the cell wall.[3] More specifically, Ag85C is important in the generation of the bacterial cell wall in that it transfers the necessary mycolic acid to the rest of the Ag85 complex, and thus it is suggested that the enzyme displays mycolyltransferace activity.[2]
Specifically, the Ag85 enzymes catalyze the transfer of a mycolyl residue from one molecule of α, α-trehalose monomycolate (TMM) to another TMM, leading to the formation of TDM (catalyze the transfer of a mycoloyl residue from one molecule of α,α-trehalose monomycolate (TMM) to another TMM, leading to the formation of TDM). Trehalose is necessary for proper growth of the bacterial cells. Thus, there is potential to target these enzymes with specific drugs to prevent the spread of the disease.[2]
Clinical Relevance
Because the cell wall envelope of M. tuberculosis is essential for the bacteria’s viability and virulence, it is the primary target for antimycobacterial drugs. More specifically, it is known that the Ag85 complex with its three protein components (A, B, and C) plays a key role in cell wall biosynthesis and the transfer of mycolic acid from one molecule of TMM to another. The resultant molecule, TDM, has been suggested to be important in maintaining M. tuberculosis cell wall integrity. Studies have also suggested that removal of Ag85C from a strain of M. tuberculosis results in a significant decrease in the presence of cell-wall linked mycolic acids. Thus, scientists hypothesize that inhibition of Ag85C and its mycolyltransferase activity will disrupt TDM and its ability to maintain cell wall integrity.[3] Knowing this, researchers have suggested that inhibition of mycolic acid transfer and, ultimately, Ag85C, must be involved in the continuously-developing class of antimycobacterial drugs aimed at combatting M. tuberculosis.[4]
Structure
Ag85 C is a dimer of identical subunits (Figure 1), and quite a bit about the amino acid sequence and overall structure of Ag85C is known. The enzyme contains a catalytic triad comprised of serine, glutamate, and histidine.
This would lead us to think that we could inhibit mycolyl transfer and shut down the enzyme by directly inhibiting the serine residue of the , but we can instead modify a near by cysteine residue and have the same effect. This was determined by a series of experiments in which scientists generated various mutants and modifications of the Ag85C enzyme.[5]
Active Site
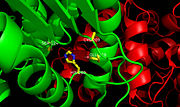
Figure 2.Ag85C with labeled active site residues. The three catalytic residues, Ser124, Glu228, and His260, as well as Cys209 are labeled.
Within the ,the homodimers shown in blue and green and the active site shown in white, three residues function together to make up the for this enzyme (Figure 2). The goal of the catalytic triad is to generate a nucleophilic residue for covalent catalysis by using an acid-base-nucleophile triad. These three residues, Ser124, Glu228, and His260 form a charge-relay network to polarize and activate the nucleophile, Ser124, which is then able to attack the substrate to form a covalent intermediate, which is then hydrolysed to regenerate a free enzyme. This charge relay is an example of the well known
chymotrypsin mechanism. Overall, it is suggested that increased enzymatic activity is attributed to the components of the active site remaining intact so that the serine nucleophile can react to form an intermediary and stabilize the transition state formed during catalysis. In the native structure, the maintains a kinked conformation necessary for correct formation of the hydrogen bonding network between the residues of the catalytic triad, thus allowing for high enzymatic activity.
[2]
Cysteine 209
is located near the , but is not directly involved in the enzyme mechanism. Recent studies suggest that this residue plays a significant role in the deactivation of Ag85C, using a mechanism not previously considered for these enzymes. Any mutation or modification to the Cys209 residue results in the relaxation of the otherwise kicked α-helix 9, which disrupts the hydrogen bonding network within the catalytic triad of the enzyme and inactivates Ag85C.[2]
Mutations and Modifications
Researchers hold particular interest in specific geometric restraints caused by ligands introduced in various Ag85C mutants and modifications: , , , and . Other mutants and modifications not described here include Ag85C-IAA, Ag85C-C209G, and Ag85C-S124A. The results demonstrate that modification of Ag85C by thiol-reactive compounds imparts structural changes in the enzyme’s active site. Specifically, scientists note a disruption in the natural kink in helix α-9. The resulting relaxed conformation of the helix significantly reduces or completely eliminates the enzymatic function of Ag85C.[2]
Ag85C-ebselen
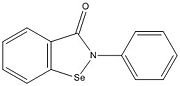
Figure 3. Ag85C-ebselen: Ebselen covalently binds to Cys209 and forces the otherwise kinked helix α-9 to take on a relaxed conformation, thus allowing movement of the helix and a reduction in enzymatic activity. The native structure is shown in blue, while the modified version is shown in purple, demonstrating a relaxed helix. Ebselen bound Cys209 is shown in green.
(Figure 3) is characterized by a covalent bond between ebselen(figure 4) and Cys209, thus forcing the otherwise kinked helix α-9 to take on a relaxed conformation (Figure 3). This allows movement of the helix and causes disruption of the hydrogen bonds within the catalytic triad, ultimately inactivating Ag85C.[2]
These initial findings suggest that ebselen-like mutants characterized by alterations in Cys209 may serve as potential drug targets for M. tuberculosis. Because any modification or mutation of Cys209 in Ag85C leads to either a dramatic decrease or complete loss of enzymatic activity, research suggests there is a low probability of M. tuberculosis developing resistance to a drug modifying the Cys209. The structures and results of the following mutations and modifications support a strategy for inhibiting the Ag85 complex as a whole with mechanism-based inhibitors that first react with Ser124 to promote the relaxation of helix α-9 and expose Cys209 and then react with Cys209 side chain thiol to . Such a bifunctional inhibitor would offer specificity while minimizing the probability of selecting for drug resistant mutants.[2]
Ag85C-Hg
Image:Ag85C Hg.jpg Figure 5. Ag85C-Hg: A modification of Ag85C generated with the addition of p-chloromercuribenzoic acid compared to the native structure. This modified enzyme lacks a hydrogen bond between Glu228 and His260, which relaxes the kink normally found in the native structure and inhibits the active site. The native structure is shown in green and the relaxed, modified structure is shown in blue.
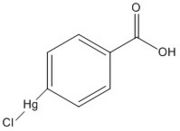
Figure 6. p-chloromercuribenzoic acid
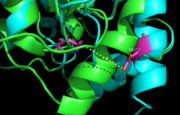
Figure 7. Ag85C-Hg active site. Glu228 and His260 are shown in red. The addition of p-chloromercuribenzoic acid disrupts the hydrogen bond between the two resides, causing a change in the helix conformation of this modified structure. The native structure is shown in green and the relaxed, modified structure is shown in blue.
The mutant Ag85C-Hg (Figure 5) is generated with the addition of p-chloromercuribenzoic acid (Figure 6), the side chain of the complex is disordered due to a lack of hydrogen bonds between Glu228 and His260. Similar to what is observed in Ag85C-ebselen, the alteration in relaxes the kinked helix α-9 found in the native structure of the enzyme, thus inhibiting the active site (Figure 7). The ultimate effect is a decrease to only 60% of the normal enzymatic function of Ag85C.[2]
Ag85C-E228Q
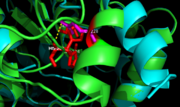
Figure 9. Ag85C-E228Q active site. The glutamate residue is shifted four angstroms in the mutated form of this enzyme causing a rearrangement of hydrogen bonds within the enzyme. The histidine residue, labeled in pink, takes on two conformations, binding with alternative serine residues, labeled in red.
The mutation introduced in Ag85C-E228Q (Figure 8) causes the Glu228 of the catalytic triad to be shifted 4 angstroms from its original position in the native structure of Ag85C (Figure 9). Due to the shift of Glu228 is the loss of hydrogen bonds between Ser124 and His260. Instead, , which also results from the shift of Glu228. A weak electron density difference in the His260 position of the native and mutated structures was also noted, suggesting that the residue may take on two alternative conformations in the mutant. Overall, the enzyme functionality is decreased to only 17% activity.[2]
Unlike other Ag85C mutants and modifications aforementioned, the does not eliminate any hydrogen bonds in the catalytic triad. Rather, it simply replaces a carboxylate moiety with an amide. Further, the structural change observed in the low-energy conformation of the mutant provides additional support for the hypothesis that the natively kinked helix α-9 is central to the enzymatic function of Ag85C.[2]
Ag85C-H260Q
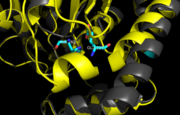
Figure 11. Ag85C-H260Q active site. A shift in helix α-9 prevents formation of any stabilizing hydrogen bonds between residues His260 and Glu228, thus decreasing its enzymatic activity.
In (Figure 10), a shift in helix α-9 between residues His260 and Glu228, thus decreasing its enzymatic activity (Figure 11). The conversion of the glutamate, a key player in the catalytic triad, to the corresponding amide-containing side chain as well as a loss of a general base in the charge relay are both key causes for the loss of function.[2]