Sandbox Reserved 1058
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
===Catalytic Loop=== | ===Catalytic Loop=== | ||
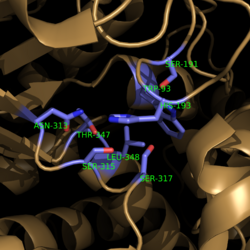
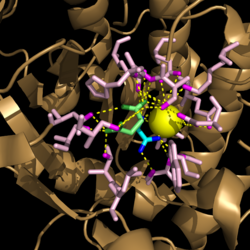
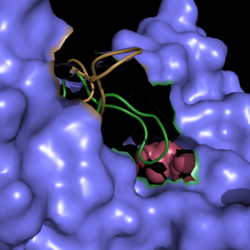
| - | [[Image:Active Loop Shift.png|250 px|left|thumb|'''Figure 4. Active Site Loop Shift.''' Binding of the ligand to the enzyme results in a conformational shift that facilitates the breakdown of isocitrate. The active site loop without a ligand bound is shown in wheat while the active site loop with a ligand bound is shown in green. The ligands are shown in raspberry.]] The <scene name='69/694225/Normal_catalytic_loop/ | + | [[Image:Active Loop Shift.png|250 px|left|thumb|'''Figure 4. Active Site Loop Shift.''' Binding of the ligand to the enzyme results in a conformational shift that facilitates the breakdown of isocitrate. The active site loop without a ligand bound is shown in wheat while the active site loop with a ligand bound is shown in green. The ligands are shown in raspberry.]] The <scene name='69/694225/Normal_catalytic_loop/4'>catalytic loop</scene> of isocitrate lyase consists of residues 185-196 ('''Figure 4'''). The two most important residues within the loop are <scene name='69/694225/Normal_catalytic_loop/3'>Cys191 and His193</scene> as these form a charge relay strong enough to extract a proton from isocitrate. Poor electron density has been observed for residues His193 and Leu194 indicating that this loop is very flexible. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> This data backs up the claim that that monomers of the protein are in a structural equilibria between the open and closed forms of the active site. In order for the catalytic loop to shift into the closed position necessary for catalysis, isocitrate must be within the binding pocket. The hydrogen bonding opportunities formed cause a ripple effect that shifts the catalytic loop into a closer position. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> This shift also causes the C-terminal domain (blue) of the subunit (residues 411-428) to <scene name='69/694225/C-terminus_loop_in_cat_loop/3'>move</scene> into the former position of the catalytic loop (green). Also shown as a reference is the ligand (pink). The C-terminal domain is then stabilized by an <scene name='69/694225/Lys_electrostatic/4'>electrostatic interaction</scene> with Lys189. This combined movement locks the active site residues into a proper orientation for lysis of a C-C bond within isocitrate. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> |
===Regulation=== | ===Regulation=== | ||
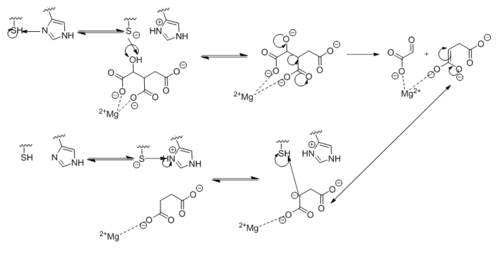
| - | <scene name='69/694225/Isocitrate_lyase/ | + | <scene name='69/694225/Isocitrate_lyase/4'>Isocitrate lyase</scene> competes with [http://en.wikipedia.org/wiki/Isocitrate_dehydrogenase isocitrate dehydrogenase], an enzyme found in the [http://en.wikipedia.org/wiki/Citric_acid_cycle citric cycle], for isocitrate processing. The favoritism of one enzyme over the other is controlled by the phosphorylation of isocitrate dehydrogenase. This enzyme has a much higher affinity for isocitrate as compared to isocitrate lyase. Phosphorylation of isocitrate dehydrogenase inactivates the enzyme and leades to increased isocitrate lyase activity. <ref name="cozzone"> Cozzone, A.; Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. ''Annual Review of Microbiology''. '''1998''', ''52'':127-164. doi: 10.1146/annurev.micro.52.1.127. </ref> |
Revision as of 17:45, 24 April 2015
Isocitrate Lyase from Mycobacterium tuberculosis
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; et. al; Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat. Struct. Biol.. 2000. 7(8):663-668.
- ↑ Gould, T.; van de Langemheen, H.; Muñoz-Elías, E.; McKinney, D.; Sacchettini, J.; Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Molecular Microbiology. 2006. 61(4):940-947. doi:10.1111/j.1365-2958.2006.05297.x.
- ↑ Cozzone, A.; Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annual Review of Microbiology. 1998, 52:127-164. doi: 10.1146/annurev.micro.52.1.127.
- ↑ Muñoz-Elías, E.; McKinney, J.; M. tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005. 11(6):638-644. doi:10.1038/nm1252.
- ↑ Srivastava, V.; Janin, A.; Srivastava, B.; Srivastava, R.; Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice. ScienceDirect. 2007. doi:10.1016/j.tube.2007.10.002.
- ↑ Dunn, M.; Ramírez-Trujillo, J.; Hernández-Lucas, I.; Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology. 2009. 155:3166-3175. doi:10.1099/mic.0.030858-0.