We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1052
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
===Ag85C-ebselen=== | ===Ag85C-ebselen=== | ||
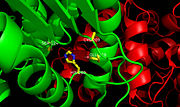
| - | [[Image:Ag85 ebselen.jpg |100 xp|left|thumb|'''Figure 3.''' [http://proteopedia.org/wiki/index.php/4qdu Ag85C-ebselen]: Ebselen covalently binds to Cys209 | + | [[Image:Ag85 ebselen.jpg |100 xp|left|thumb|'''Figure 3.''' [http://proteopedia.org/wiki/index.php/4qdu Ag85C-ebselen]: Ebselen covalently binds to Cys209 and forces the otherwise kinked helix α-9 to take on a relaxed conformation, thus allowing movement of the helix and a reduction in enzymatic activity. The native structure is shown in blue while the modified version is shown in purple, demonstrating a relaxed helix. Ebselen-bound Cys209 is shown in green.]] |
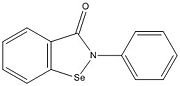
| - | [[Image:ebselen pic.jpg |100 xp| | + | [[Image:ebselen pic.jpg |100 xp|right|thumb|'''Figure 4.''' Ebselen.]] |
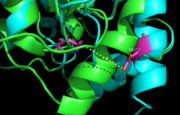
<scene name='69/694218/Ebselen/1'>Ag85C-Ebselen</scene> (Figure 3) is characterized by a covalent bond between [http://en.wikipedia.org/wiki/Ebselen ebselen](figure 4) and Cys209, thus forcing the otherwise kinked helix α-9 to take on a relaxed conformation (Figure 3). This allows movement of the helix and causes disruption of the hydrogen bonds within the catalytic triad, ultimately inactivating Ag85C.<ref name="Favrot"/> | <scene name='69/694218/Ebselen/1'>Ag85C-Ebselen</scene> (Figure 3) is characterized by a covalent bond between [http://en.wikipedia.org/wiki/Ebselen ebselen](figure 4) and Cys209, thus forcing the otherwise kinked helix α-9 to take on a relaxed conformation (Figure 3). This allows movement of the helix and causes disruption of the hydrogen bonds within the catalytic triad, ultimately inactivating Ag85C.<ref name="Favrot"/> | ||
These initial findings suggest that ebselen-like mutants characterized by alterations in Cys209 may serve as potential drug targets for ''M. tuberculosis''. Because any modification or mutation of Cys209 in Ag85C leads to either a dramatic decrease or complete loss of enzymatic activity, research suggests there is a low probability of ''M. tuberculosis'' developing resistance to a drug modifying the Cys209. The structures and results of the following mutations and modifications support a strategy for inhibiting the Ag85 complex as a whole with mechanism-based inhibitors that first react with Ser124 to promote the relaxation of helix α-9 and expose Cys209 and then react with Cys209 side chain thiol to <scene name='69/694218/Ebselen/2'>covalently modify this conserved residue</scene>. Such a bifunctional inhibitor would offer specificity while minimizing the probability of selecting for drug resistant mutants.<ref name="Favrot"/> | These initial findings suggest that ebselen-like mutants characterized by alterations in Cys209 may serve as potential drug targets for ''M. tuberculosis''. Because any modification or mutation of Cys209 in Ag85C leads to either a dramatic decrease or complete loss of enzymatic activity, research suggests there is a low probability of ''M. tuberculosis'' developing resistance to a drug modifying the Cys209. The structures and results of the following mutations and modifications support a strategy for inhibiting the Ag85 complex as a whole with mechanism-based inhibitors that first react with Ser124 to promote the relaxation of helix α-9 and expose Cys209 and then react with Cys209 side chain thiol to <scene name='69/694218/Ebselen/2'>covalently modify this conserved residue</scene>. Such a bifunctional inhibitor would offer specificity while minimizing the probability of selecting for drug resistant mutants.<ref name="Favrot"/> | ||
| + | |||
| + | {{clear}} | ||
===Ag85C-Hg=== | ===Ag85C-Hg=== | ||
| - | [[Image:Ag85C Hg.jpg |100 xp|left|thumb|'''Figure 5.''' [http://proteopedia.org/wiki/index.php/4qdo Ag85C-Hg]: A modification of Ag85C generated with the addition of p-chloromercuribenzoic acid compared to the native structure. This modified enzyme lacks a hydrogen bond between Glu228 and His260, which relaxes the kink normally found in the native structure and inhibits the active site. The native structure is shown in green and the relaxed, modified structure is shown in blue.]] | ||
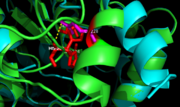
| - | [[Image: | + | [[Image:ag85c Hg zoom.png|100 xp|left|thumb|'''Figure 5.''' [http://proteopedia.org/wiki/index.php/4qdo Ag85C-Hg] active site. Glu228 and His260 are shown in red. The addition of p-chloromercuribenzoic acid disrupts the hydrogen bond between the two resides, causing a change in the helix conformation of this modified structure. The native structure is shown in green and the relaxed, modified structure is shown in blue.]] |
| - | [[Image:ag85c Hg zoom.png|100 xp|left|thumb|'''Figure 7.''' [http://proteopedia.org/wiki/index.php/4qdo Ag85C-Hg] active site. Glu228 and His260 are shown in red. The addition of p-chloromercuribenzoic acid disrupts the hydrogen bond between the two resides, causing a change in the helix conformation of this modified structure. The native structure is shown in green and the relaxed, modified structure is shown in blue.]] | ||
| + | [[Image:P-chloromercuribenzoic acid.jpg |100 xp|right|thumb|'''Figure 6.''' p-chloromercuribenzoic acid]] | ||
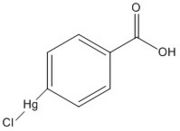
| - | The mutant [http://proteopedia.org/wiki/index.php/4qdo Ag85C-Hg] | + | The mutant [http://proteopedia.org/wiki/index.php/4qdo Ag85C-Hg] is generated with the addition of [http://en.wikipedia.org/wiki/4-Chloromercuribenzoic_acid p-chloromercuribenzoic acid] (Figure 6), the side chain of the complex is disordered due to a lack of hydrogen bonds between Glu228 and His260. Similar to what is observed in Ag85C-ebselen, the alteration in <scene name='69/694218/4qdo/1'>Ag85C-Hg</scene> relaxes the kinked helix α-9 found in the native structure of the enzyme, thus inhibiting the active site (Figure 5). The ultimate effect is a decrease to only 60% of the normal enzymatic function of Ag85C.<ref name="Favrot"/> |
| + | |||
| + | {{clear}} | ||
===Ag85C-E228Q=== | ===Ag85C-E228Q=== | ||
| - | [[Image:E228Q.jpg |100 xp|left|thumb|'''Figure 8.''' [http://proteopedia.org/wiki/index.php/4qdz Ag85C-E228Q].]] | ||
| - | [[Image:E228Q zoom.png |100 xp|left|thumb|'''Figure 9.''' [http://proteopedia.org/wiki/index.php/4qdz Ag85C-E228Q] active site. The glutamate residue is shifted four angstroms in the mutated form of this enzyme causing a rearrangement of hydrogen bonds within the enzyme. The histidine residue, labeled in pink, takes on two conformations, binding with alternative serine residues, labeled in red.]] | ||
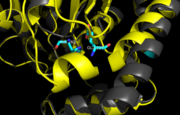
| - | The mutation introduced in [http://proteopedia.org/wiki/index.php/4qdz Ag85C-E228Q] | + | [[Image:E228Q zoom.png |100 xp|left|thumb|'''Figure 7.''' [http://proteopedia.org/wiki/index.php/4qdz Ag85C-E228Q] active site. The glutamate residue is shifted four angstroms in the mutated form of this enzyme causing a rearrangement of hydrogen bonds within the enzyme. The histidine residue, labeled in pink, takes on two conformations, binding with alternative serine residues, labeled in red.]] |
| + | |||
| + | The mutation introduced in [http://proteopedia.org/wiki/index.php/4qdz Ag85C-E228Q]causes the Glu228 of the catalytic triad to be shifted 4 angstroms from its original position in the native structure of Ag85C (Figure 7). Due to the shift of Glu228 is the loss of hydrogen bonds between Ser124 and His260. Instead, <scene name='69/694218/4qdz/2'>His260 bonds with Ser148</scene>, which also results from the shift of Glu228. A weak electron density difference in the His260 position of the native and mutated structures was also noted, suggesting that the residue may take on two alternative conformations in the <scene name='69/694218/Ag85c-e228q/1'>Ag85C-E228Q</scene> mutant. Overall, the enzyme functionality is decreased to only 17% activity.<ref name="Favrot"/> | ||
Unlike other Ag85C mutants and modifications aforementioned, the <scene name='69/694218/Ag85c-e228q/1'>Ag85C-E228Q</scene>does not eliminate any hydrogen bonds in the catalytic triad. Rather, it simply replaces a carboxylate moiety with an amide. Further, the structural change observed in the low-energy conformation of the <scene name='69/694218/Ag85c-e228q/1'>Ag85C-E228Q</scene> mutant provides additional support for the hypothesis that the natively kinked helix α-9 is central to the enzymatic function of Ag85C.<ref name="Favrot"/> | Unlike other Ag85C mutants and modifications aforementioned, the <scene name='69/694218/Ag85c-e228q/1'>Ag85C-E228Q</scene>does not eliminate any hydrogen bonds in the catalytic triad. Rather, it simply replaces a carboxylate moiety with an amide. Further, the structural change observed in the low-energy conformation of the <scene name='69/694218/Ag85c-e228q/1'>Ag85C-E228Q</scene> mutant provides additional support for the hypothesis that the natively kinked helix α-9 is central to the enzymatic function of Ag85C.<ref name="Favrot"/> | ||
| + | |||
| + | {{clear}} | ||
===Ag85C-H260Q=== | ===Ag85C-H260Q=== | ||
| - | [[Image:H260Q.jpg |100 xp|left|thumb|'''Figure 10.''' [http://proteopedia.org/wiki/index.php/4qe3 Ag85C-H260Q]: Mutated Ag85C enzyme due to conversion of Glu228 to amide-containing side chain.]] | ||
| - | [[Image:H260Q zoom.png|100 xp|left|thumb|'''Figure | + | [[Image:H260Q zoom.png|100 xp|left|thumb|'''Figure 8.''' [http://proteopedia.org/wiki/index.php/4qe3 Ag85C-H260Q] active site. A shift in helix α-9 prevents formation of any stabilizing hydrogen bonds between residues His260 and Glu228, thus decreasing its enzymatic activity.]] |
| - | In <scene name='69/694218/Mutationh260q/1'>Ag85C-H260Q</scene> | + | In <scene name='69/694218/Mutationh260q/1'>Ag85C-H260Q</scene>, a shift in helix α-9 <scene name='69/694218/Ag85c-hg/1'>prevents the hydrogen bond formation</scene>between residues His260 and Glu228, thus decreasing its enzymatic activity (Figure 8). The conversion of the glutamate, a key player in the catalytic triad, to the corresponding amide-containing side chain as well as a loss of a general base in the charge relay are both key causes for the loss of function.<ref name="Favrot"/> |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 18:02, 25 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Ag85C of Mycobacterium tuberculosis
| |||||||||||
References
- ↑ Online Mendelian Inheritance in Man (OMIM). Mycobacterium tuberculosis, susceptibility to. [Internet]. Baltimore (MD): Johns Hopkins University; 2014 Nov 12 [cited 2015 March 16]. Available from http://www.omim.org/entry/607948
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 Favrot L, Lajiness DH, Ronning DR. Inactivation of the Mycobacterium tuberculosis Antigen 85 complex by covalent, allosteric inhibitors. J Biol Chem. 2014 Jul 14. pii: jbc.M114.582445. PMID:25028518 doi:http://dx.doi.org/10.1074/jbc.M114.582445
- ↑ 3.0 3.1 Gobec S, Plantan I, Mravljak J, Wilson RA, Besra GS, Kikelj D. Phosphonate inhibitors of antigen 85C, a crucial enzyme involved in the biosynthesis of the Mycobacterium tuberculosis cell wall. Bioorg Med Chem Lett. 2004 Jul 5;14(13):3559-62. PMID:15177473 doi:http://dx.doi.org/10.1016/j.bmcl.2004.04.052
- ↑ Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997 May 30;276(5317):1420-2. PMID:9162010
- ↑ Research Collaboratory for Structural Bioinformatics Protein Database (RCSB PDB) Diacylglycerol acyltransferase/mycolyltransferase Ag85C - P9WQN9 (A85C_MYCTU). [Internet] San Diego (CA): University of San Diego; 2014 [cited 2015 March 16]. DOI:10.2210/pdbp9wqn9/pdb