We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Cytochrome c
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
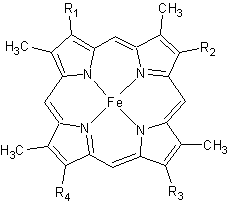
[[Image:heme.gif |frame|left| Figure 2. The tetrapyrrolic heme prosthetic group that can either be covalently attached to or closely associated with various proteins, such as cytochromes and other globin proteins. In ''Rm''cyt''c'', R2 is an ethyl group covalently attached to Cys 45, and R3 is a methyl group covalently attached to Cys48.]] | [[Image:heme.gif |frame|left| Figure 2. The tetrapyrrolic heme prosthetic group that can either be covalently attached to or closely associated with various proteins, such as cytochromes and other globin proteins. In ''Rm''cyt''c'', R2 is an ethyl group covalently attached to Cys 45, and R3 is a methyl group covalently attached to Cys48.]] | ||
| - | + | {{Clear}} | |
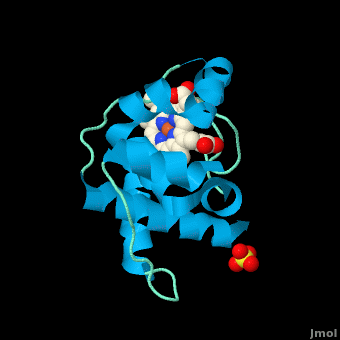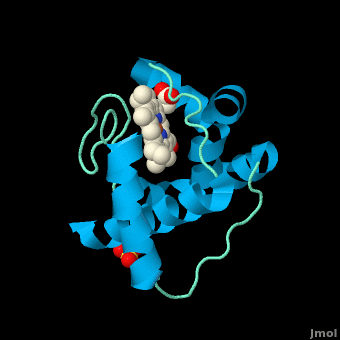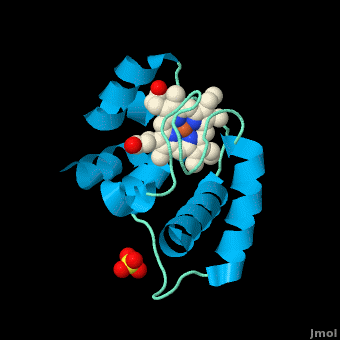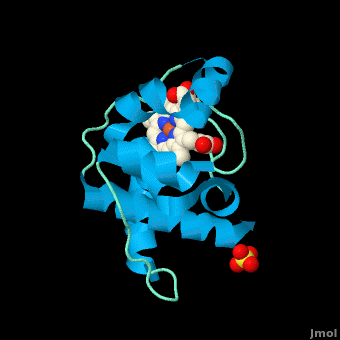
The typical monoheme cyt ''c'' fold is formed by helices <scene name='Sandbox_Reserved_335/Helices/4'>A, C, and E</scene>. ''Rm''cyt''c'' contains seven α-helices that are folded around the heme, all connected by random coils.<ref name=main /> The heme group is axially coordinated by <scene name='Sandbox_Reserved_335/Axial/6'>His49 and Met100</scene>, and the disulfide linkages exist at <scene name='Sandbox_Reserved_335/Cys/1'>Cys45 and Cys48</scene>. The heme group in ''Rm''cyt''c'' is almost completely shielded from solvent due to it being in a mostly hydrophobic pocket. This pocket is formed in part by the seven helices surrounding the ring, but also by two structures that are uncommon in other cytochromes ''c''. First, a 21 amino acid extension of the N-terminal exists, forming <scene name='Sandbox_Reserved_335/Uncommon1/2'>α-helix A' and loop 1</scene>, which wraps around the back of the polypeptide.<ref name=main /> An extension resembling such has only been seen in ''Thermus thermophilus''; however, the extension occurs at the C-terminus rather than the N-terminus.<ref>doi:10.1006/jmbi.1997.1181</ref> A second rarity is that of <scene name='Sandbox_Reserved_335/Uncommon2/2'>helix B'</scene>, inserted between helix D and loop 3, that shields the bottom part of the heme from any solvent.<ref name=main /> In cytochrome ''c''<sub>2</sub> as well as mitochondrial cyt ''c'', a similar yet shorter helix was found, though this helix was present at a different place in the primary sequence. Also, instead of helix B', ''T. thermophilus'' contains a two-stranded [http://en.wikipedia.org/wiki/Beta_sheet β-sheet].<ref name=main /> One final note is the number of <scene name='Sandbox_Reserved_335/Met/1'>methionine</scene> residues that ''Rm''cyt''c'' contains. In general, cyt ''c'' contains about two methionines whereas ''Rm''cyt''c'' contains seven, located on the left of the heme.<ref name=main /> | The typical monoheme cyt ''c'' fold is formed by helices <scene name='Sandbox_Reserved_335/Helices/4'>A, C, and E</scene>. ''Rm''cyt''c'' contains seven α-helices that are folded around the heme, all connected by random coils.<ref name=main /> The heme group is axially coordinated by <scene name='Sandbox_Reserved_335/Axial/6'>His49 and Met100</scene>, and the disulfide linkages exist at <scene name='Sandbox_Reserved_335/Cys/1'>Cys45 and Cys48</scene>. The heme group in ''Rm''cyt''c'' is almost completely shielded from solvent due to it being in a mostly hydrophobic pocket. This pocket is formed in part by the seven helices surrounding the ring, but also by two structures that are uncommon in other cytochromes ''c''. First, a 21 amino acid extension of the N-terminal exists, forming <scene name='Sandbox_Reserved_335/Uncommon1/2'>α-helix A' and loop 1</scene>, which wraps around the back of the polypeptide.<ref name=main /> An extension resembling such has only been seen in ''Thermus thermophilus''; however, the extension occurs at the C-terminus rather than the N-terminus.<ref>doi:10.1006/jmbi.1997.1181</ref> A second rarity is that of <scene name='Sandbox_Reserved_335/Uncommon2/2'>helix B'</scene>, inserted between helix D and loop 3, that shields the bottom part of the heme from any solvent.<ref name=main /> In cytochrome ''c''<sub>2</sub> as well as mitochondrial cyt ''c'', a similar yet shorter helix was found, though this helix was present at a different place in the primary sequence. Also, instead of helix B', ''T. thermophilus'' contains a two-stranded [http://en.wikipedia.org/wiki/Beta_sheet β-sheet].<ref name=main /> One final note is the number of <scene name='Sandbox_Reserved_335/Met/1'>methionine</scene> residues that ''Rm''cyt''c'' contains. In general, cyt ''c'' contains about two methionines whereas ''Rm''cyt''c'' contains seven, located on the left of the heme.<ref name=main /> | ||
| Line 31: | Line 31: | ||
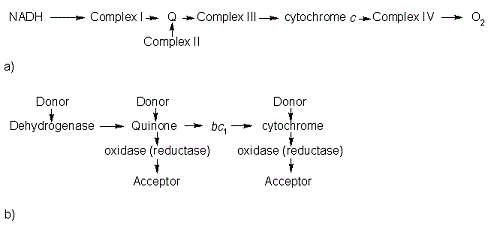
In the electron transport chain (ETC), cyt ''c'' shuttles electrons between the respiratory complexes III and IV; complex III is the cytochrome ''bc''<sub>1</sub> complex and IV is cyt ''c'' oxidase. Initially, the heme iron in cyt ''c'' is in the reduced, Fe<sup>3+</sup> state; this allows for the uptake of one electron, oxidizing the iron to the Fe<sup>2+</sup> state.<ref name='etc'>Karp, Gerald (2008). Cell and Molecular Biology (5th edition). Hoboken, NJ: John Wiley & Sons. ISBN 978-0470042175.</ref> The ETC in eukaryotes is quite simple compared to that of prokaryotes (Figure 3). | In the electron transport chain (ETC), cyt ''c'' shuttles electrons between the respiratory complexes III and IV; complex III is the cytochrome ''bc''<sub>1</sub> complex and IV is cyt ''c'' oxidase. Initially, the heme iron in cyt ''c'' is in the reduced, Fe<sup>3+</sup> state; this allows for the uptake of one electron, oxidizing the iron to the Fe<sup>2+</sup> state.<ref name='etc'>Karp, Gerald (2008). Cell and Molecular Biology (5th edition). Hoboken, NJ: John Wiley & Sons. ISBN 978-0470042175.</ref> The ETC in eukaryotes is quite simple compared to that of prokaryotes (Figure 3). | ||
| - | [[Image:Etc.gif |frame|left| | | + | [[Image:Etc.gif |frame|left|thumb|400px| Figure 3. The electron transport chain of a) eukaryotes as compared to b) prokaryotes.]] |
In prokaryotic systems, electrons can enter the ETC at a number of places and multiple donors can be in play; however, the underlying transport system remains the same. Electrons are ultimately transferred from donor to various redox complexes including the ''bc''<sub>1</sub> complex and cytochrome ''c'', and finally to a terminal electron acceptor such as molecular oxygen in eukaryotes.<ref name=etc /> | In prokaryotic systems, electrons can enter the ETC at a number of places and multiple donors can be in play; however, the underlying transport system remains the same. Electrons are ultimately transferred from donor to various redox complexes including the ''bc''<sub>1</sub> complex and cytochrome ''c'', and finally to a terminal electron acceptor such as molecular oxygen in eukaryotes.<ref name=etc /> | ||
Revision as of 12:55, 29 April 2015
| |||||||||||
3D structures of cytochrome C
Updated on 29-April-2015
References
- ↑ Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001 Nov 2;313(4):903-19. PMID:11697912 doi:10.1006/jmbi.2001.5080
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 Stelter M, Melo AM, Pereira MM, Gomes CM, Hreggvidsson GO, Hjorleifsdottir S, Saraiva LM, Teixeira M, Archer M. A Novel Type of Monoheme Cytochrome c: Biochemical and Structural Characterization at 1.23 A Resolution of Rhodothermus marinus Cytochrome c. Biochemistry. 2008 Oct 15. PMID:18855424 doi:10.1021/bi800999g
- ↑ 3.0 3.1 3.2 Reedy CJ, Gibney BR. Heme protein assemblies. Chem Rev. 2004 Feb;104(2):617-49. PMID:14871137 doi:10.1021/cr0206115
- ↑ 4.0 4.1 4.2 Ambler RP. Sequence variability in bacterial cytochromes c. Biochim Biophys Acta. 1991 May 23;1058(1):42-7. PMID:1646017
- ↑ Cookson DJ, Moore GR, Pitt RC, Williams RJP, Campbell ID, Ambler RP, Bruschi M, Le Gall J. Structural homology of cytochromes c. Eur J Biochem. 1978 Feb;83(1):261-75.
- ↑ Than ME, Hof P, Huber R, Bourenkov GP, Bartunik HD, Buse G, Soulimane T. Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing. J Mol Biol. 1997 Aug 29;271(4):629-44. PMID:9281430 doi:10.1006/jmbi.1997.1181
- ↑ Soares CM, Baptista AM, Pereira MM, Teixeira M. Investigation of protonatable residues in Rhodothermus marinus caa3 haem-copper oxygen reductase: comparison with Paracoccus denitrificans aa3 haem-copper oxygen reductase. J Biol Inorg Chem. 2004 Mar;9(2):124-34. Epub 2003 Dec 23. PMID:14691678 doi:10.1007/s00775-003-0509-9
- ↑ Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001 Jun 1;1505(2-3):185-208. PMID:11334784
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 Karp, Gerald (2008). Cell and Molecular Biology (5th edition). Hoboken, NJ: John Wiley & Sons. ISBN 978-0470042175.
- ↑ Rajagopal BS, Wilson MT, Bendall DS, Howe CJ, Worrall JA. Structural and kinetic studies of imidazole binding to two members of the cytochrome c (6) family reveal an important role for a conserved heme pocket residue. J Biol Inorg Chem. 2011 Jan 26. PMID:21267610 doi:10.1007/s00775-011-0758-y
- ↑ Morelli X, Czjzek M, Hatchikian CE, Bornet O, Fontecilla-Camps JC, Palma NP, Moura JJ, Guerlesquin F. Structural model of the Fe-hydrogenase/cytochrome c553 complex combining transverse relaxation-optimized spectroscopy experiments and soft docking calculations. J Biol Chem. 2000 Jul 28;275(30):23204-10. PMID:10748163 doi:10.1074/jbc.M909835199
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, Melissa Morrison, Adis Hasic