We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1065
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
= Introduction = | = Introduction = | ||
| - | ''Mycobacterium tuberculosis'' very-long-chain fatty acyl-CoA synthetase, also known as <scene name='69/694233/General_pic/1'>FadD13</scene>, is unique within its class of FadD proteins in regards to its ability to house [https://en.wikipedia.org/wiki/Lipid lipid] substrates longer than itself. These lipid substrates are very-long-chain fatty acids between lengths C22 –C26, which is up to the maximum length tested. <ref name="Our Paper"/> The significance of | + | ''Mycobacterium tuberculosis'' very-long-chain fatty acyl-CoA synthetase, also known as <scene name='69/694233/General_pic/1'>FadD13</scene>, is unique within its class of FadD proteins in regards to its ability to house [https://en.wikipedia.org/wiki/Lipid lipid] substrates longer than itself. These lipid substrates are very-long-chain fatty acids between lengths C22 –C26, which is up to the maximum length tested. <ref name="Our Paper"/> The significance of these very-long-chain fatty acids lies in their importance to mycolic acid synthesis by ''Mycobacterium tuberculosis'' ''(M. tb)''. Mycolic acids compose part of the cell wall of ''(M. tb)''. FadD13, an activator of mycolic acids, has been identified as key component in the virulence of [https://en.wikipedia.org/wiki/Mycobacterium_tuberculosis ''Mycobacterium tuberculosis''], the etiological agent of [https://en.wikipedia.org/wiki/Tuberculosis tuberculosis], and has emerged as possible target for novel therapeutic agents.<ref name="JT">PMID: 20454815</ref> The FadD13 enzyme is the last gene of the ''mymA'' operon. <ref name="residue paper"/> |
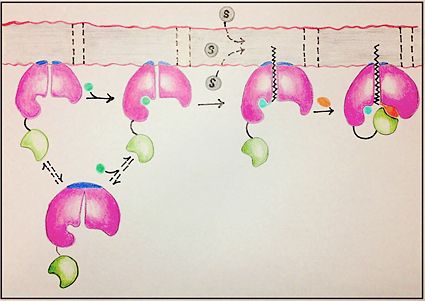
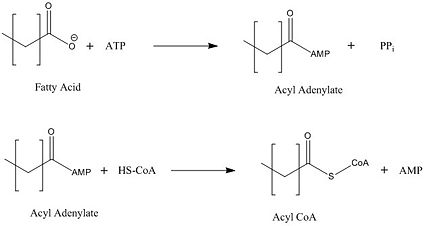
There are four main groups of FadD enzymes categorized on their ability to accommodate different length substrates: short (C2-C4), medium (C4-C12), long (C12-C22), and very long (C22-C26).<ref name="Our Paper"/> Most FadD class proteins exist as integral membrane proteins, involved in both the activation of fatty acids and other hydrophobic substrates in addition to the transport of these lipids into the cell. However, substrates longer than the enzyme itself, like these very-long-chain fatty acids, pose an interesting structural dilemma to the enzyme. FadD13 differs from typical integral membrane fatty acyl-CoA synthetases in that FadD13 exists as a [https://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral membrane protein]. This key feature provides a mechanistic basis for FadD13’s activation and transport of fatty acids of length C24-C26 through the two step addition of [http://en.wikipedia.org/wiki/Coenzyme_A Coenzyme A](Figure 1).<ref name="Our Paper">PMID: 22560731</ref> | There are four main groups of FadD enzymes categorized on their ability to accommodate different length substrates: short (C2-C4), medium (C4-C12), long (C12-C22), and very long (C22-C26).<ref name="Our Paper"/> Most FadD class proteins exist as integral membrane proteins, involved in both the activation of fatty acids and other hydrophobic substrates in addition to the transport of these lipids into the cell. However, substrates longer than the enzyme itself, like these very-long-chain fatty acids, pose an interesting structural dilemma to the enzyme. FadD13 differs from typical integral membrane fatty acyl-CoA synthetases in that FadD13 exists as a [https://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral membrane protein]. This key feature provides a mechanistic basis for FadD13’s activation and transport of fatty acids of length C24-C26 through the two step addition of [http://en.wikipedia.org/wiki/Coenzyme_A Coenzyme A](Figure 1).<ref name="Our Paper">PMID: 22560731</ref> | ||
Revision as of 15:26, 1 May 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Very-Long-Chain Fatty Acyl-CoA Synthetase
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Andersson CS, Lundgren CA, Magnusdottir A, Ge C, Wieslander A, Molina DM, Hogbom M. The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme. Structure. 2012 May 2. PMID:22560731 doi:10.1016/j.str.2012.03.012
- ↑ Jatana N, Jangid S, Khare G, Tyagi AK, Latha N. Molecular modeling studies of Fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis--a potential target for the development of antitubercular drugs. J Mol Model. 2011 Feb;17(2):301-13. doi: 10.1007/s00894-010-0727-3. Epub 2010 May, 8. PMID:20454815 doi:http://dx.doi.org/10.1007/s00894-010-0727-3
- ↑ 3.0 3.1 3.2 3.3 3.4 Khare G, Gupta V, Gupta RK, Gupta R, Bhat R, Tyagi AK. Dissecting the role of critical residues and substrate preference of a Fatty Acyl-CoA Synthetase (FadD13) of Mycobacterium tuberculosis. PLoS One. 2009 Dec 21;4(12):e8387. doi: 10.1371/journal.pone.0008387. PMID:20027301 doi:10.1371/journal.pone.0008387
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Jatana N, Jangid S, Khare G, Tyagi AK, Latha N. Molecular modeling studies of Fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis--a potential target for the development of antitubercular drugs. J Mol Model. 2011 Feb;17(2):301-13. doi: 10.1007/s00894-010-0727-3. Epub 2010 May, 8. PMID:20454815 doi:http://dx.doi.org/10.1007/s00894-010-0727-3
- ↑ 5.0 5.1 5.2 Schroeder EK, de Souza N, Santos DS, Blanchard JS, Basso LA. Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis. Curr Pharm Biotechnol. 2002 Sep;3(3):197-225. PMID:12164478
Similar Proteopedia Pages
Phosphopantetheinyl Transferase
External Resources
Lipids Wikipedia page
Tuberculosis Wikipedia page
Mycobacterium tuberculosis Wikipedia page
Coenzyme A Wikipedia page
Acyl CoA Wikipedia Page
Mycolic Acid Wikipedia page
Peripheral Membrane Protein Wikipedia page
Ethionamide Wikipedia page
Isoniazid Wikipedia page
Thiocarlide Wikipedia page
Pyrazinamide Wikipedia page