We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 977
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
== Structural highlights == | == Structural highlights == | ||
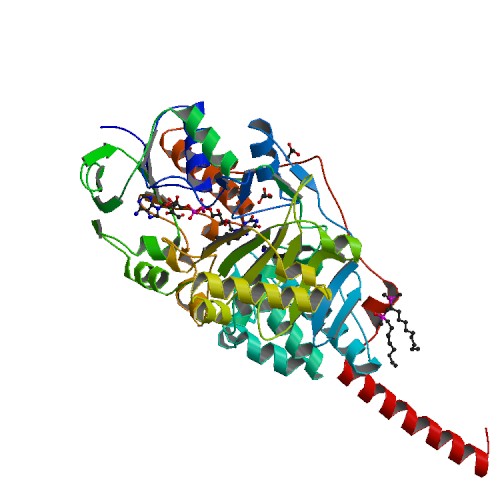
MAOA has multiple binding flavin adenine dinucleotide (FAD)-binding domains, and multiple substrate-binding sites. A, C-terminal alpha-helical region and hydrophobic residues 17, 18, and 21 bind to the mitochondrial membrane. Aromatic and aliphatic residues make up most of the substrate-binding sites. The lysine305 residue interacts with a water molecule and binds flavin cofactors. Tyr407 tyr444 help to form an "aromatic sandwich" that help orient the substrate for activation by promoting nucleophicity. The binding site is about 400Å in size. Two residues contribute to substate selectivity and inhibitor selectivity are Ile335 and Phe208. There are three tyrosine, Tyr69,Tyr407, Tyr444, near the active sites. <ref>Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.</ref> Overall, the active site area is outlined by residues Tyr69, Tyr197, Phe208, Tyr407, Phe352, Tyr444, and the isoalloxazine ring. <ref>E. Jones, T., Giurato, L., Guccione, S., & R. Ramsay, R. (2007). Interactions of Imidazoline Ligands with the Active Site of Purified Monoamine Oxidase A. The FEBS Journal, 1567-1575. Retrieved January 1, 2015, from Scifinder.</ref> | MAOA has multiple binding flavin adenine dinucleotide (FAD)-binding domains, and multiple substrate-binding sites. A, C-terminal alpha-helical region and hydrophobic residues 17, 18, and 21 bind to the mitochondrial membrane. Aromatic and aliphatic residues make up most of the substrate-binding sites. The lysine305 residue interacts with a water molecule and binds flavin cofactors. Tyr407 tyr444 help to form an "aromatic sandwich" that help orient the substrate for activation by promoting nucleophicity. The binding site is about 400Å in size. Two residues contribute to substate selectivity and inhibitor selectivity are Ile335 and Phe208. There are three tyrosine, Tyr69,Tyr407, Tyr444, near the active sites. <ref>Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.</ref> Overall, the active site area is outlined by residues Tyr69, Tyr197, Phe208, Tyr407, Phe352, Tyr444, and the isoalloxazine ring. <ref>E. Jones, T., Giurato, L., Guccione, S., & R. Ramsay, R. (2007). Interactions of Imidazoline Ligands with the Active Site of Purified Monoamine Oxidase A. The FEBS Journal, 1567-1575. Retrieved January 1, 2015, from Scifinder.</ref> | ||
| - | + | <scene name='70/700893/Catalytic_site/1'>TextToBeDisplayed</scene> | |
| - | + | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 14:37, 4 May 2015
Monoamine Oxidase A (MAOA)
| |||||||||||
References
- ↑ Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.
- ↑ Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.
- ↑ Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.
- ↑ Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.
- ↑ Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.
- ↑ E. Jones, T., Giurato, L., Guccione, S., & R. Ramsay, R. (2007). Interactions of Imidazoline Ligands with the Active Site of Purified Monoamine Oxidase A. The FEBS Journal, 1567-1575. Retrieved January 1, 2015, from Scifinder.