This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox 977
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Function == | == Function == | ||
| - | The major function of this enzyme is to degrade biogenic amines. MAOAs catalyze the oxidation of serotonin, norepinephrine, and dopamine. Then hydrolyze the oxidative products to give aldehydes or ketones. <ref>Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.</ref | + | The major function of this enzyme is to degrade biogenic amines. MAOAs catalyze the oxidation of serotonin, norepinephrine, and dopamine. Then hydrolyze the oxidative products to give aldehydes or ketones. <ref>Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.</ref> |
== Disease == | == Disease == | ||
MAOAs remove amino functional groups to leave an oxidized oxygen atom behind, and producing ammonia and hydrogen peroxide. The hydrogen peroxide is a major mediator in the production of hydroxyl radicals in the body. Free hydroxyl radicals in the body have detrimental effects on several organs especially the brain. MAOAs are associated with major depressive disorder and cardiovascular disease. Increased activity of this enzyme can lead to downstream dysregulations of elevation of oxidative stresses, increased platelet activity, and cytokine levels.<ref>Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.</ref> | MAOAs remove amino functional groups to leave an oxidized oxygen atom behind, and producing ammonia and hydrogen peroxide. The hydrogen peroxide is a major mediator in the production of hydroxyl radicals in the body. Free hydroxyl radicals in the body have detrimental effects on several organs especially the brain. MAOAs are associated with major depressive disorder and cardiovascular disease. Increased activity of this enzyme can lead to downstream dysregulations of elevation of oxidative stresses, increased platelet activity, and cytokine levels.<ref>Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.</ref> | ||
| Line 11: | Line 11: | ||
It is important to study MAOAs because they are so harmful to the human body. Many of these effects can be avoided with the use of monoamine oxidase A inhibitors. These inhibitors block the substrate binding sites, keeping free amines from binding and being metabolized by the MAOAs. .<ref>Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.</ref> | It is important to study MAOAs because they are so harmful to the human body. Many of these effects can be avoided with the use of monoamine oxidase A inhibitors. These inhibitors block the substrate binding sites, keeping free amines from binding and being metabolized by the MAOAs. .<ref>Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.</ref> | ||
== Structural highlights == | == Structural highlights == | ||
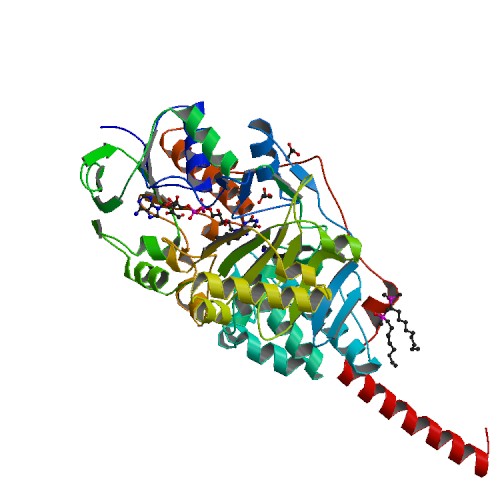
| - | MAOA has multiple binding flavin adenine dinucleotide (FAD)-binding domains, and multiple substrate-binding sites. A, C-terminal alpha-helical region and hydrophobic residues 17, 18, and 21 bind to the mitochondrial membrane. Aromatic and aliphatic residues make up most of the substrate-binding sites. The lysine305 residue interacts with a water molecule and binds flavin cofactors. Tyr407 tyr444 help to form an "aromatic sandwich" that help orient the substrate for activation by promoting nucleophicity. The binding site is about 400Å in size. Two residues contribute to substate selectivity and inhibitor selectivity are Ile335 and Phe208. There are three tyrosine, Tyr69,Tyr407, Tyr444, near the active sites. <ref>Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.</ref> Overall, the active site area is outlined by residues Tyr69, Tyr197, Phe208, Tyr407, Phe352, Tyr444, and the isoalloxazine ring. <ref>E. Jones, T., Giurato, L., Guccione, S., & R. Ramsay, R. (2007). Interactions of Imidazoline Ligands with the Active Site of Purified Monoamine Oxidase A. The FEBS Journal, 1567-1575. Retrieved January 1, 2015, from Scifinder.</ref> | + | MAOA has multiple binding flavin adenine dinucleotide (FAD)-binding domains, and multiple substrate-binding sites. A, C-terminal alpha-helical region and hydrophobic residues 17, 18, and 21 bind to the mitochondrial membrane. Aromatic and aliphatic residues make up most of the substrate-binding sites. The lysine305 residue interacts with a water molecule and binds flavin cofactors. Tyr407 tyr444 help to form an "aromatic sandwich" that help orient the substrate for activation by promoting nucleophicity. The binding site is about 400Å in size. Two residues contribute to substate selectivity and inhibitor selectivity are Ile335 and Phe208. There are three tyrosine, Tyr69,Tyr407, Tyr444, near the active sites. <ref>Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.</ref> Overall, the active site area is outlined by residues Tyr69, Tyr197, Phe208, Tyr407, Phe352, Tyr444, and the isoalloxazine ring. <ref>E. Jones, T., Giurato, L., Guccione, S., & R. Ramsay, R. (2007). Interactions of Imidazoline Ligands with the Active Site of Purified Monoamine Oxidase A. The FEBS Journal, 1567-1575. Retrieved January 1, 2015, from Scifinder.</ref><scene name='70/700893/Catalytic_site/2'>The active site of MAOA with the amino groups highlighted in green.</scene> |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
Monoamine Oxidase A (MAOA)
| |||||||||||
References
- ↑ Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.
- ↑ Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.
- ↑ Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.
- ↑ Machado-Vieira, R., & G. Mallinger, A. (2012). Abnormal Function of Monoamine Oxidase-A in comorbid Major Depressive disorder and Cardiovascular Disease: Pathophysical and Theraputic Implications (Review). Molecular Medicine Reports, 6, 915-922. Retrieved April 2, 2015, from Scifinder.
- ↑ Gaweska, Helena. "Structures and Mechanism of the Monoamine Oxidase Family." BioMol Concepts 2 (2011): 365-77. Print.
- ↑ E. Jones, T., Giurato, L., Guccione, S., & R. Ramsay, R. (2007). Interactions of Imidazoline Ligands with the Active Site of Purified Monoamine Oxidase A. The FEBS Journal, 1567-1575. Retrieved January 1, 2015, from Scifinder.