Regulator of G protein signaling
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
<imagemap> | <imagemap> | ||
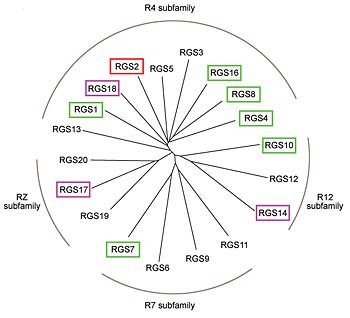
Image:Kosloff-NSMB2011-Fig1b.jpg|350px|family of canonical RGS proteins | Image:Kosloff-NSMB2011-Fig1b.jpg|350px|family of canonical RGS proteins | ||
| - | <ref>PMID: 21685921</ref> | ||
poly 131 45 213 41 210 110 127 109 [[2ace]] | poly 131 45 213 41 210 110 127 109 [[2ace]] | ||
</imagemap> | </imagemap> | ||
| - | + | <ref>PMID: 21685921</ref> | |
| - | + | ||
In these proteins, the ~120-residue RGS homology domain functions as a GTPase-activating protein (GAP) for GTP-bound Gα subunits. In addition to these domains, diverse proteins subfamilies that include the ~120-residue RGS homology domain bear additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity. | In these proteins, the ~120-residue RGS homology domain functions as a GTPase-activating protein (GAP) for GTP-bound Gα subunits. In addition to these domains, diverse proteins subfamilies that include the ~120-residue RGS homology domain bear additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity. | ||
Revision as of 12:07, 13 May 2015
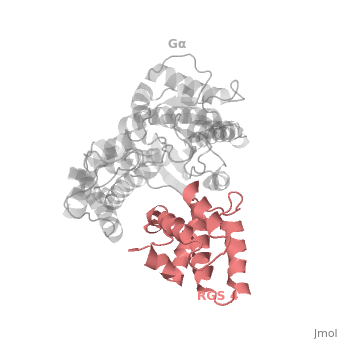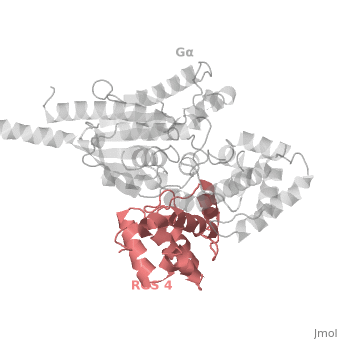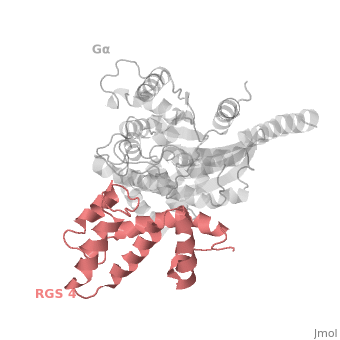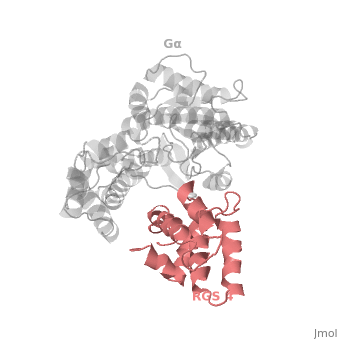
Regulator of G protein signaling (RGS) interactions with G proteins – RGS4-Gαi as a model structure.
| |||||||||||
References
- ↑ Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY. Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions. Nat Struct Mol Biol. 2011 Jun 19;18(7):846-53. doi: 10.1038/nsmb.2068. PMID:21685921 doi:http://dx.doi.org/10.1038/nsmb.2068
- ↑ Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997 Apr 18;89(2):251-61. PMID:9108480
Proteopedia Page Contributors and Editors (what is this?)
Ali Asli, Denise Salem, Michal Harel, Joel L. Sussman, Jaime Prilusky