Regulator of G protein signaling
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
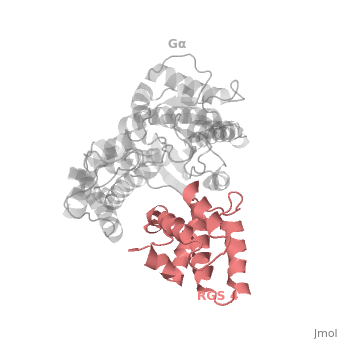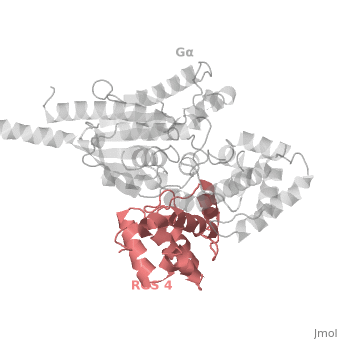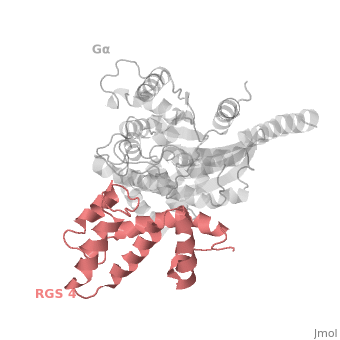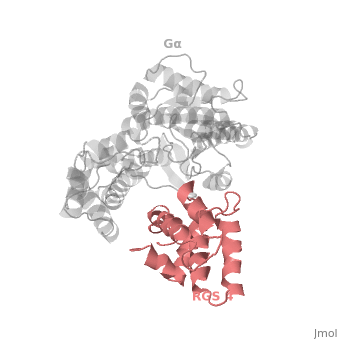
RGS proteins are selective for binding to the transition state of Gα(GTP → GDP + P<sub>i</sub>), which can be mimicked by Gα-GDP bound with the planar ion aluminum tetrafluoride (AlF<sub>4−</sub>). | RGS proteins are selective for binding to the transition state of Gα(GTP → GDP + P<sub>i</sub>), which can be mimicked by Gα-GDP bound with the planar ion aluminum tetrafluoride (AlF<sub>4−</sub>). | ||
| - | Like many signaling proteins, RGS proteins comprise a large and diverse family. In human genome, Thirty-seven RGS proteins are encoded by gene loci; this collection of related proteins has been divided into 10 different subfamilies based on the relatedness of their RGS domain sequence and their multiple domain architectures. About 20 ‘canonical’ RGS proteins can in theory downregulate any of the 16 activated Gα subunits, although in practice they interact only with members of the G<sub>i</sub> and G<sub>q</sub> families. | + | Like many signaling proteins, RGS proteins comprise a large and diverse family. In human genome, Thirty-seven RGS proteins are encoded by gene loci; this collection of related proteins has been divided into 10 different subfamilies based on the relatedness of their RGS domain sequence and their multiple domain architectures. About 20 ‘canonical’ RGS proteins can in theory downregulate any of the 16 activated Gα subunits, although in practice they interact only with members of the G<sub>i</sub> and G<sub>q</sub> families. |
| + | In these proteins, the ~120-residue RGS homology domain functions as a GTPase-activating protein (GAP) for GTP-bound Gα subunits. | ||
<imagemap> | <imagemap> | ||
| Line 11: | Line 12: | ||
poly 131 45 213 41 210 110 127 109 [[2ace]] | poly 131 45 213 41 210 110 127 109 [[2ace]] | ||
</imagemap> | </imagemap> | ||
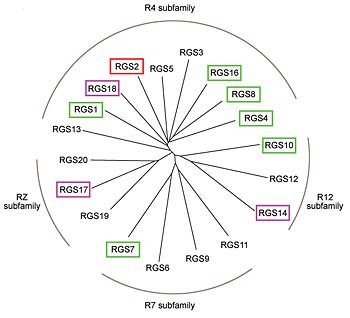
| + | Phylogenetic tree of 19 human RGS domains. | ||
| + | RGS proteins whose activity was tested are colored. In green RGS proteins with high GAP activity, in purple RGS proteins with low but discernible activities and in red, RGS2 had no measurable activity. | ||
<ref>PMID: 21685921</ref> | <ref>PMID: 21685921</ref> | ||
| - | + | ||
| + | In addition to these domains, diverse proteins subfamilies that include the ~120-residue RGS homology domain bear additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity. | ||
== Heterotrimeric G-proteins family == | == Heterotrimeric G-proteins family == | ||
Revision as of 12:16, 13 May 2015
Regulator of G protein signaling (RGS) interactions with G proteins – RGS4-Gαi as a model structure.
| |||||||||||
References
- ↑ Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY. Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions. Nat Struct Mol Biol. 2011 Jun 19;18(7):846-53. doi: 10.1038/nsmb.2068. PMID:21685921 doi:http://dx.doi.org/10.1038/nsmb.2068
- ↑ Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997 Apr 18;89(2):251-61. PMID:9108480
Proteopedia Page Contributors and Editors (what is this?)
Ali Asli, Denise Salem, Michal Harel, Joel L. Sussman, Jaime Prilusky