Sandbox Reserved 1051
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
==Biological Role== | ==Biological Role== | ||
| - | The unusually thick and waxy cell wall of [http://en.wikipedia.org/wiki/Mycobacterium mycobacteria] is primarily composed of [http://en.wikipedia.org/wiki/Peptidoglycan peptidoglycans], [http://en.wikipedia.org/wiki/Arabinogalactan arbinogalactans], and [http://en.wikipedia.org/wiki/Mycolic_acid mycolic acids]. The mycolic acids, which have very long carbon chains and exhibit extreme hydrophobicity, are responsible for forming the outermost layer of the cell wall, thus creating a hydrophobic envelope surrounding the mycobacterium. Much of the mycolic acid content of the cell wall is in the form of esters (trehalose-6-monomycolate, TMM) and bis-esters (trehalose-6,6'-dimycolate, TDM, cord factor) of trehalose. | + | ===Cell Wall of Mycobacteria=== |
| - | + | The unusually thick and waxy cell wall of [http://en.wikipedia.org/wiki/Mycobacterium mycobacteria] is primarily composed of [http://en.wikipedia.org/wiki/Peptidoglycan peptidoglycans], [http://en.wikipedia.org/wiki/Arabinogalactan arbinogalactans], and [http://en.wikipedia.org/wiki/Mycolic_acid mycolic acids]. The mycolic acids, which have very long carbon chains and exhibit extreme hydrophobicity, are responsible for forming the outermost layer of the cell wall, thus creating a hydrophobic envelope surrounding the mycobacterium. Much of the mycolic acid content of the cell wall is in the form of esters (trehalose-6-monomycolate, TMM) and bis-esters (trehalose-6,6'-dimycolate, TDM, [http://en.wikipedia.org/wiki/Cord_factor cord factor]) of [http://en.wikipedia.org/wiki/Trehalose trehalose]. | |
| - | The antigen 85 (Ag85) complex in [http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis ''Mycobacterium tuberculosis''] is composed of three homologous proteins: Ag85A, Ag85B, and Ag85C, encoded by the ''fbpA'', ''fbpB'', and ''fbpC'' genes, respectively. Each of these Ag85 components (A, B, and C) is an acyltransferase enzyme (EC 2.3.1.122) that can catalyze mycolyl transfer from | + | ===Enzymatic Activity=== |
| + | The antigen 85 (Ag85) complex in [http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis ''Mycobacterium tuberculosis''] is composed of three homologous proteins: Ag85A, Ag85B, and Ag85C, encoded by the ''fbpA'', ''fbpB'', and ''fbpC'' genes, respectively. Each of these Ag85 components (A, B, and C) is an acyltransferase enzyme (EC 2.3.1.122) that can catalyze mycolyl transfer from TMM to another alcoholic substrate, including TMM itself (Figure 1). | ||
NEED REFERENCES! | NEED REFERENCES! | ||
INSERT '''SIMPLE''' REACTION SCHEME AS FIGURE 1 (see Gobec et al.) | INSERT '''SIMPLE''' REACTION SCHEME AS FIGURE 1 (see Gobec et al.) | ||
| Line 20: | Line 21: | ||
==Structure== | ==Structure== | ||
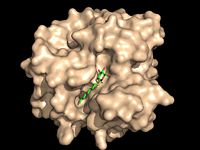
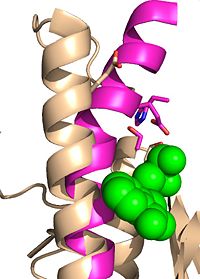
| - | [[Image:Substrate Binding 2D Surface.jpg|200 px|left|thumb|'''Figure 2:''' Octylthioglucoside, a substrate analog, shown in the binding pocket of [ | + | [[Image:Substrate Binding 2D Surface.jpg|200 px|left|thumb|'''Figure 2:''' Octylthioglucoside (green), a substrate analog, shown in the binding pocket of Ag85C (tan, from [[1va5]])]] |
| - | Wild type Ag85C, in the absence of any specific ligands, was originally crystallized as a <scene name='69/694219/Dimer/1'>homodimer</scene>.<ref name="Ronning2000">PMID: 10655617</ref> The <scene name='69/694220/Secondary_structures/2'>secondary structure</scene> (shown in the monomeric form) is 38% α-helical (pink) and 16% β-sheet (yellow). The confrontation of a central β-sheet bordered by α–helices comprises an [http://en.wikipedia.org/wiki/Alpha/beta_hydrolase_fold α/β hydrolase fold] in Ag85C, a supersecondary structure that is highly conserved across serine hydrolases. The active site of Ag85C is composed of two adjoined binding pockets; there is carbohydrate binding pocket for the trehalose substrate, and there is a fatty acid binding pocket for the mycolic acid. As a result, trehalose monomycolate can effectively bind to the Ag85C active site. This binding pocket is shown in Figure 2, in which Ag85C is shown with a bound substrate mimic, octylthioglucoside (Figure 3). | + | Wild type Ag85C, in the absence of any specific ligands, was originally crystallized as a <scene name='69/694219/Dimer/1'>homodimer</scene>.<ref name="Ronning2000">PMID: 10655617</ref> The <scene name='69/694220/Secondary_structures/2'>secondary structure</scene> (shown in the monomeric form) is 38% α-helical (pink) and 16% β-sheet (yellow). The confrontation of a central β-sheet bordered by α–helices comprises an [http://en.wikipedia.org/wiki/Alpha/beta_hydrolase_fold α/β hydrolase fold] in Ag85C, a supersecondary structure that is highly conserved across serine hydrolases. The active site of Ag85C is composed of two adjoined binding pockets; there is carbohydrate binding pocket for the trehalose substrate, and there is a fatty acid binding pocket for the mycolic acid. As a result, trehalose monomycolate can effectively bind to the Ag85C active site. This binding pocket is shown in Figure 2, in which Ag85C is shown with a bound substrate mimic, octylthioglucoside<ref name="Ronning2004">PMID: 15192106</ref> (Figure 3). |
| + | |||
| + | INSERT CHEMDRAW PICTURE COMPARING THE OCTYLTHIOGLYCOSIDE STRUCTURE TO TMM | ||
| - | == | + | ===Catalytic Triad=== |
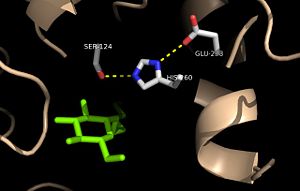
[[Image:Substrate_Catalytic_Triad.jpg|300 px|right|thumb|'''Figure 2:''' Relation of the catalytic triad to the octylthioglucoside analog in [http://www.rcsb.org/pdb/explore/explore.do?structureId=1VA5 Ag85C]]] | [[Image:Substrate_Catalytic_Triad.jpg|300 px|right|thumb|'''Figure 2:''' Relation of the catalytic triad to the octylthioglucoside analog in [http://www.rcsb.org/pdb/explore/explore.do?structureId=1VA5 Ag85C]]] | ||
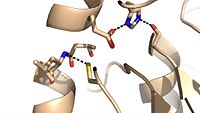
| - | Mutagenesis studies have confirmed the Ag85C functions through a Glu-His-Ser <scene name='69/694220/Catalytic_triad/4'>catalytic triad</scene>, similar to that of [http://en.wikipedia.org/wiki/Chymotrypsin chymotrypsin]. By modifying each of the catalytic residues separately testing the enzyme’s relative activity, it has been shown that mutation of any one of these residues dramatically reduces activity. The S124 alcohol’s nucleophilicity is inductively strengthened through H260 and E224, which allows the S124 residue to catalyze a reaction that involves [http://en.wikipedia.org/wiki/Cord_factor trehalose 6, 6’-dimycolate]. The formation of the functional catalytic triad relies on upon Van der Waals interaction between C209 and the peptide bond between L232 and T231. This interaction results in a kinked conformation of the α9 helix, which promotes that activity of the catalytic triad. As a result, Ag85C, a mycolyl transferase, can facilitate the modification of trehalose monomycolates to trehalose dimycolates, which are then transported to the bacterial cell wall. This reaction is shown in '''Figure 3''' below. | + | Mutagenesis studies<ref name="Favrot2014"> PMID:25028518 </ref> have confirmed the Ag85C functions through a Glu-His-Ser <scene name='69/694220/Catalytic_triad/4'>catalytic triad</scene>, similar to that of [http://en.wikipedia.org/wiki/Chymotrypsin chymotrypsin]. By modifying each of the catalytic residues separately testing the enzyme’s relative activity, it has been shown that mutation of any one of these residues dramatically reduces activity. The S124 alcohol’s nucleophilicity is inductively strengthened through H260 and E224, which allows the S124 residue to catalyze a reaction that involves [http://en.wikipedia.org/wiki/Cord_factor trehalose 6, 6’-dimycolate]. The formation of the functional catalytic triad relies on upon Van der Waals interaction between C209 and the peptide bond between L232 and T231. This interaction results in a kinked conformation of the α9 helix, which promotes that activity of the catalytic triad. As a result, Ag85C, a mycolyl transferase, can facilitate the modification of trehalose monomycolates to trehalose dimycolates, which are then transported to the bacterial cell wall. This reaction is shown in '''Figure 3''' below. |
[[Image:Mech_Ag85C.jpeg|400 px|center|thumb|'''Figure 3:''' General reaction catalyzed by Antigen 85C]] | [[Image:Mech_Ag85C.jpeg|400 px|center|thumb|'''Figure 3:''' General reaction catalyzed by Antigen 85C]] | ||
Revision as of 19:07, 13 May 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Trehalose-O-mycolyltransferase Ag85C
Introduction
Antigen 85C is one of three homologous protein components of the Ag85 complex in the cell wall of M. tuberculosis. This serine esterase enzyme catalyzes the transfer of mycolyl groups, characteristic components of the cell wall of mycobacteria. Several three dimensional structures of Ag85C have been solved, including the wild type enzyme as well as active site variants due to site-directed mutagenesis and covalent modification.
| |||||||||||
References
- ↑ Jackson M, Raynaud C, Laneelle MA, Guilhot C, Laurent-Winter C, Ensergueix D, Gicquel B, Daffe M. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol Microbiol. 1999 Mar;31(5):1573-87. PMID:10200974
- ↑ Ronning DR, Klabunde T, Besra GS, Vissa VD, Belisle JT, Sacchettini JC. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat Struct Biol. 2000 Feb;7(2):141-6. PMID:10655617 doi:10.1038/72413
- ↑ Ronning DR, Vissa V, Besra GS, Belisle JT, Sacchettini JC. Mycobacterium tuberculosis antigen 85A and 85C structures confirm binding orientation and conserved substrate specificity. J Biol Chem. 2004 Aug 27;279(35):36771-7. Epub 2004 Jun 10. PMID:15192106 doi:http://dx.doi.org/10.1074/jbc.M400811200
- ↑ Favrot L, Lajiness DH, Ronning DR. Inactivation of the Mycobacterium tuberculosis Antigen 85 complex by covalent, allosteric inhibitors. J Biol Chem. 2014 Jul 14. pii: jbc.M114.582445. PMID:25028518 doi:http://dx.doi.org/10.1074/jbc.M114.582445