Regulator of G protein signaling
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
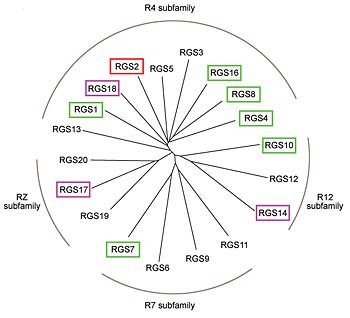
RGS proteins whose activity was tested are colored by their GAP activity, RGS proteins with high GAP activity (green), RGS proteins with low but discernible activities (purple) and RGS2 had no measurable activity (red). | RGS proteins whose activity was tested are colored by their GAP activity, RGS proteins with high GAP activity (green), RGS proteins with low but discernible activities (purple) and RGS2 had no measurable activity (red). | ||
<ref>PMID: 21685921</ref> | <ref>PMID: 21685921</ref> | ||
| - | In addition to these domains, diverse proteins subfamilies that include the ~120-residue RGS homology domain bear additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity. R7-subfamily members share a multi-domain protein architecture composed of DEP and GGL domains on the N-terminal side of the RGS domain. R12-subfamily members possess a tandem repeat of Ras binding domains (RBDs) and a single GoLoco motif. | + | In addition to these domains, diverse proteins subfamilies that include the ~120-residue RGS homology domain bear additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity. R7-subfamily members share a multi-domain protein architecture composed of DEP and GGL domains on the N-terminal side of the RGS domain. R12-subfamily members possess a tandem repeat of Ras binding domains (RBDs) and a single GoLoco motif. |
== Heterotrimeric G-proteins family == | == Heterotrimeric G-proteins family == | ||
| Line 22: | Line 22: | ||
== Structural highlights == | == Structural highlights == | ||
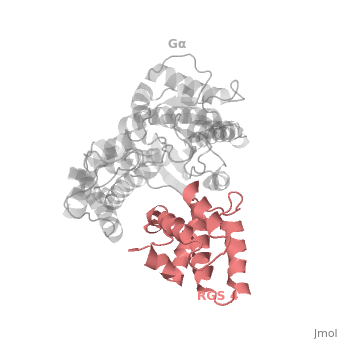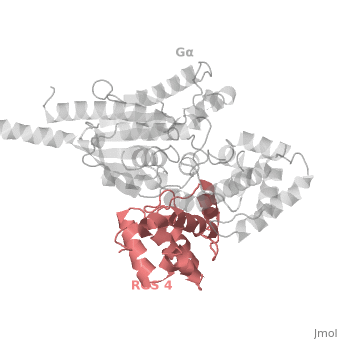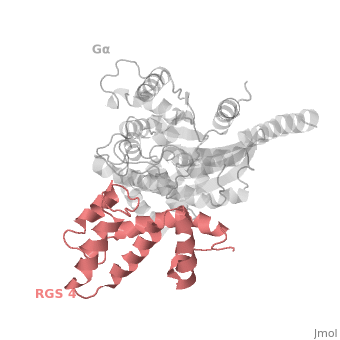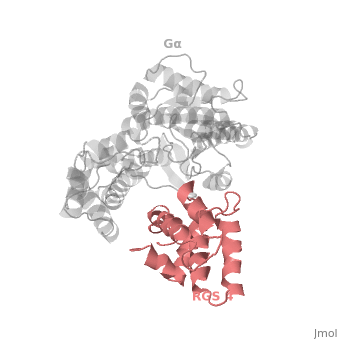
| - | The structure of the RGS domain was defined by X-ray crystallographic analysis of a complex of RGS4 and Gαi1. | + | The structure of the RGS domain was defined by X-ray crystallographic analysis of a complex of RGS4 and Gαi1. |
1AGR is a tetramer structure of two identical duplicate crystal complex of <scene name='70/701447/Gi-rgs4/3'>RGS4- Gα<sub>i1</sub></scene> (tetramer excess stability of crystal structure). | 1AGR is a tetramer structure of two identical duplicate crystal complex of <scene name='70/701447/Gi-rgs4/3'>RGS4- Gα<sub>i1</sub></scene> (tetramer excess stability of crystal structure). | ||
<scene name='70/701447/Rgs_monomer/11'>Monomer structure of RGS4</scene> in darkmagenta cartoon diagram: The RGS4 domain corresponds to an array of nine α-helices that fold into two small subdomains. The terminal subdomain contains the N and C termini of the box and is formed by α1, α2, α3, α8, and α9. Helices α1 and α9 lie in antiparallel orientation, juxtaposing the N and C termini of the box. The larger bundle subdomain, formed by α4, α5, α6, and α7, is a classic right-handed, antiparallel four-helix bundle. Both subdomains are required for GAP activity. | <scene name='70/701447/Rgs_monomer/11'>Monomer structure of RGS4</scene> in darkmagenta cartoon diagram: The RGS4 domain corresponds to an array of nine α-helices that fold into two small subdomains. The terminal subdomain contains the N and C termini of the box and is formed by α1, α2, α3, α8, and α9. Helices α1 and α9 lie in antiparallel orientation, juxtaposing the N and C termini of the box. The larger bundle subdomain, formed by α4, α5, α6, and α7, is a classic right-handed, antiparallel four-helix bundle. Both subdomains are required for GAP activity. | ||
Revision as of 07:27, 14 May 2015
Regulator of G protein signaling (RGS) interactions with G proteins – RGS4-Gαi as a model structure.
| |||||||||||
References
- ↑ Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY. Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions. Nat Struct Mol Biol. 2011 Jun 19;18(7):846-53. doi: 10.1038/nsmb.2068. PMID:21685921 doi:http://dx.doi.org/10.1038/nsmb.2068
- ↑ Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997 Apr 18;89(2):251-61. PMID:9108480
Proteopedia Page Contributors and Editors (what is this?)
Ali Asli, Denise Salem, Michal Harel, Joel L. Sussman, Jaime Prilusky