This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Connexin
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
[[Image:distances b.jpg]] | [[Image:distances b.jpg]] | ||
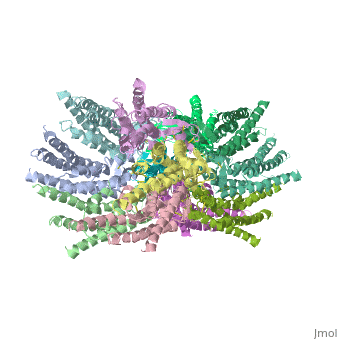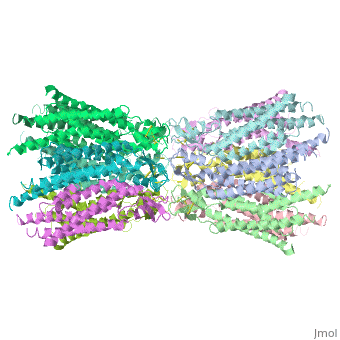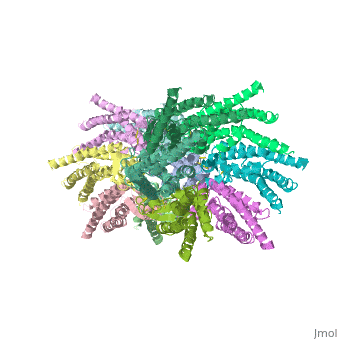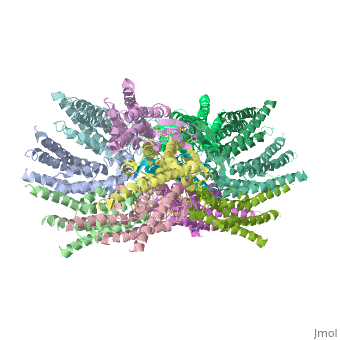
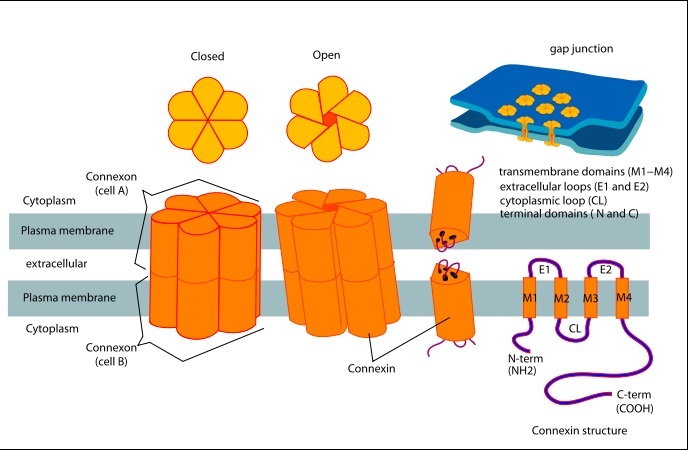
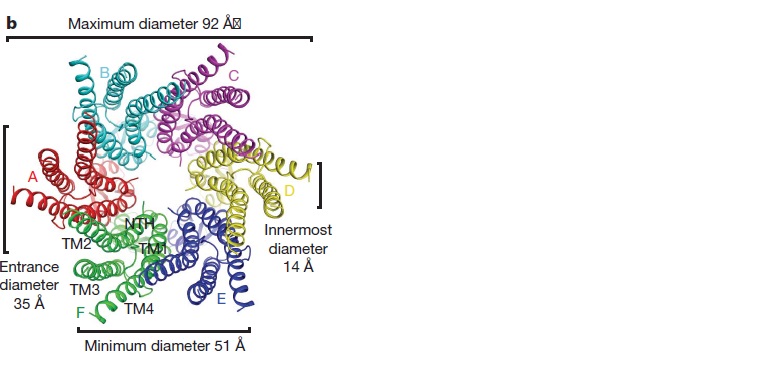
==Structure of the cx26 protomer:== | ==Structure of the cx26 protomer:== | ||
| - | The protomer has four transmembrane segments (TM1–4), two extracellular loops (E1 and E2), a cytoplasmic loop, an N-terminal helix (NTH), and a C-terminal segment (Fig. 3). Cx26 forms a typical four-helix bundle in which any pair of adjacent helices is antiparallel. TM1 and TM2 face the interior, whereas TM3 and TM4 face the hydrophobic membrane environment. There has been controversy about the identity of the major pore-lining helix, on the basis of accessibility studies of substituted cysteines and sequence analysis. One set of data favours TM3 as the major pore helix and the other favours TM1 . The helical arrangement of our structure is consistent with the latter model. The major pore-lining helix TM1 is inclined, so that the pore diameter narrows from the cytoplasmic to the extracellular side of the membrane, and ends in a short 310 helix. | + | The [[http://en.wikipedia.org/wiki/Promoter_(genetics) protomer]] has four transmembrane segments (TM1–4), two extracellular loops (E1 and E2), a cytoplasmic loop, an N-terminal helix (NTH), and a C-terminal segment (Fig. 3). Cx26 forms a typical four-helix bundle in which any pair of adjacent helices is antiparallel. TM1 and TM2 face the interior, whereas TM3 and TM4 face the hydrophobic membrane environment. There has been controversy about the identity of the major pore-lining helix, on the basis of accessibility studies of substituted cysteines and sequence analysis. One set of data favours TM3 as the major pore helix and the other favours TM1 . The helical arrangement of our structure is consistent with the latter model. The major pore-lining helix TM1 is inclined, so that the pore diameter narrows from the cytoplasmic to the extracellular side of the membrane, and ends in a short 310 helix. |
The extracellular loop E1 contains a 310 helix at the beginning and a short a-helix in its C-terminal half E2, together with E1, contains a short antiparallel b-sheet and | The extracellular loop E1 contains a 310 helix at the beginning and a short a-helix in its C-terminal half E2, together with E1, contains a short antiparallel b-sheet and | ||
stretches over E1, forming the outside wall of the connexon. Six conserved cysteine residues, three in each loop, form intramolecular disulphide bonds between E1 and E2 Most of the prominent intra-protomer interactions are in the extracellular part of the transmembrane region, Our structure revealed the interactions between the two adjoining connexons of the gap junction channel, which involve both E1 and E2 . The N-terminal half of E2 seems rather flexible | stretches over E1, forming the outside wall of the connexon. Six conserved cysteine residues, three in each loop, form intramolecular disulphide bonds between E1 and E2 Most of the prominent intra-protomer interactions are in the extracellular part of the transmembrane region, Our structure revealed the interactions between the two adjoining connexons of the gap junction channel, which involve both E1 and E2 . The N-terminal half of E2 seems rather flexible | ||
| Line 24: | Line 24: | ||
==Pore funnel and the voltage-dependent gating mechanism:== | ==Pore funnel and the voltage-dependent gating mechanism:== | ||
| - | The short NTHs of the six protomers formthe funnel , This finding agrees with an NMR solution structure of an N-terminal peptide of Cx26, which showed that the loop connecting the NTH to TM1 is very | + | The short NTHs of the six protomers formthe funnel , This finding agrees with an [[http://chem.ch.huji.ac.il/nmr/whatisnmr/whatisnmr.html NMR]] solution structure of an N-terminal peptide of Cx26, which showed that the loop connecting the NTH to TM1 is very flexible. Asp 2 forms hydrogen bonds with the main chain amide of Thr 5 from the neighbouring protomer. The Asp 2 and Thr 5 residues on neighbouring NTHs at the bottom of the funnel form a circular girdle, as previously seen in the nicotinic acetylcholine receptor, which stabilizes the funnel structure . <ref name='Structure'/> |
=Differences between wild type and mutant connexin 26:= | =Differences between wild type and mutant connexin 26:= | ||
In general, single site mutations are spread fairly evenly across the whole protein with TM2 having the highest mutation density (number of amino acids with NHLS mutations divided by the total number of amino acids in the domain) at 67% to M1 and E1 having the lowest density of mutations with their respective domains at 33%. According to this criterion, TM4 has a mutation density of 40%. . Of the four transmembrane helices, M1, M2 and M3 have attracted the most attention, because of the controversies involved in models with different helix assignments, based on lower resolution cryo-electron crystallographic structures and scanning cysteine accessibility mutagenesis . Far less is known about TM4 and how side chains interact with the other helices and with the lipid bilayer. <ref name='mutant int'/> | In general, single site mutations are spread fairly evenly across the whole protein with TM2 having the highest mutation density (number of amino acids with NHLS mutations divided by the total number of amino acids in the domain) at 67% to M1 and E1 having the lowest density of mutations with their respective domains at 33%. According to this criterion, TM4 has a mutation density of 40%. . Of the four transmembrane helices, M1, M2 and M3 have attracted the most attention, because of the controversies involved in models with different helix assignments, based on lower resolution cryo-electron crystallographic structures and scanning cysteine accessibility mutagenesis . Far less is known about TM4 and how side chains interact with the other helices and with the lipid bilayer. <ref name='mutant int'/> | ||
Revision as of 12:06, 17 May 2015
| |||||||||||
References
- ↑ Zonta F, Buratto D, Cassini C, Bortolozzi M, Mammano F. Molecular dynamics simulations highlight structural and functional alterations in deafness-related M34T mutation of connexin 26. Front Physiol. 2014 Mar 4;5:85. doi: 10.3389/fphys.2014.00085. eCollection 2014. PMID:24624091 doi:http://dx.doi.org/10.3389/fphys.2014.00085
- ↑ 2.0 2.1 2.2 2.3 2.4 Suga M, Maeda S, Nakagawa S, Yamashita E, Tsukihara T. A description of the structural determination procedures of a gap junction channel at 3.5 A resolution. Acta Crystallogr D Biol Crystallogr. 2009 Aug;65(Pt 8):758-66. Epub 2009, Jul 10. PMID:19622859 doi:http://dx.doi.org/10.1107/S0907444909014711
- ↑ http://en.wikipedia.org/wiki/Connexin
- ↑ 4.0 4.1 Ambrosi C, Walker AE, Depriest AD, Cone AC, Lu C, Badger J, Skerrett IM, Sosinsky GE. Analysis of trafficking, stability and function of human connexin 26 gap junction channels with deafness-causing mutations in the fourth transmembrane helix. PLoS One. 2013 Aug 15;8(8):e70916. doi: 10.1371/journal.pone.0070916. eCollection, 2013. PMID:23967136 doi:http://dx.doi.org/10.1371/journal.pone.0070916
- ↑ 5.0 5.1 5.2 Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y. Asymmetric Configurations and N-terminal Rearrangements in Connexin26 Gap Junction Channels. J Mol Biol. 2011 Jan 21;405(3):724-35. Epub 2010 Nov 20. PMID:21094651 doi:10.1016/j.jmb.2010.10.032
Proteopedia Page Contributors and Editors (what is this?)
Safaa Salah Hussiesy, Michal Harel, Doaa Naffaa, Jaime Prilusky