This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1051
From Proteopedia
| Line 36: | Line 36: | ||
==Structures of Variant Enzymes== | ==Structures of Variant Enzymes== | ||
===Covalent Modification of Cys209=== | ===Covalent Modification of Cys209=== | ||
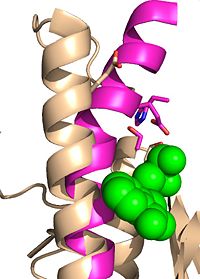
| - | Ag85C has been crystallized following covalent inhibition by several thiol-modifying agents: ebselen, ''p''-chloromercuribenzoic acid, and iodoacetamide. Each of these thiol-reactive inhibitors covalently bound to C209 and caused a relaxation of the α9 helix. The resulting relaxed conformation of the helix significantly reduces or completely eliminates the enzymatic function of Ag85C.<ref name=" | + | Ag85C has been crystallized following covalent inhibition by several thiol-modifying agents: ebselen, ''p''-chloromercuribenzoic acid, and iodoacetamide. Each of these thiol-reactive inhibitors covalently bound to C209 and caused a relaxation of the α9 helix. The resulting relaxed conformation of the helix significantly reduces or completely eliminates the enzymatic function of Ag85C.<ref name="Favrot2014"/> |
====Modification by Ebselen==== | ====Modification by Ebselen==== | ||
| Line 44: | Line 44: | ||
====Modification by ''p''-Chloromercuribenzoic acid==== | ====Modification by ''p''-Chloromercuribenzoic acid==== | ||
| - | The thiol of C209 can also be covalently modified by [http://en.wikipedia.org/wiki/4-Chloromercuribenzoic_acid p-chloromercuribenzoic acid]. Similar to what is observed in Ag85C-ebselen, the alteration in <scene name='69/694218/4qdo/1'>Ag85C-Hg</scene> relaxes the kinked helix α-9 found in the native structure of the enzyme. The native structure is shown in green and the relaxed, modified structure is shown in blue. This conformational change again disrupts the hydrogen bonding network of the catalytic triad, resulting in a decrease to only 60% of the normal enzymatic function<ref name=" | + | The thiol of C209 can also be covalently modified by [http://en.wikipedia.org/wiki/4-Chloromercuribenzoic_acid p-chloromercuribenzoic acid]. Similar to what is observed in Ag85C-ebselen, the alteration in <scene name='69/694218/4qdo/1'>Ag85C-Hg</scene> relaxes the kinked helix α-9 found in the native structure of the enzyme. The native structure is shown in green and the relaxed, modified structure is shown in blue. This conformational change again disrupts the hydrogen bonding network of the catalytic triad, resulting in a decrease to only 60% of the normal enzymatic function<ref name="Favrot2014"/>. |
===Mutation of Cys209=== | ===Mutation of Cys209=== | ||
====E228Q==== | ====E228Q==== | ||
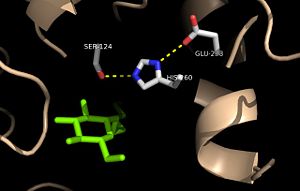
| - | <scene name='69/694218/Ag85c-e228q/1'>Mutation of the glutamate component of the catalytic triad, E228, to glutamine</scene> decreased enzymatic activity to only 17% of the wild type<ref name=" | + | <scene name='69/694218/Ag85c-e228q/1'>Mutation of the glutamate component of the catalytic triad, E228, to glutamine</scene> decreased enzymatic activity to only 17% of the wild type<ref name="Favrot2014"/>. The α-9 helix is again relaxed in this variant. The mutated residue was shifted 4 angstroms from its original position in the native structure. Associated with this shift of Glu228 is the loss of hydrogen bonds between Ser124 and His260. The His260 residue takes on two conformations, <scene name='69/694218/4qdz/2'>in which it can hydrogen bond to either Ser124 or Ser148</scene>. |
{{clear}} | {{clear}} | ||
| Line 55: | Line 55: | ||
====H260Q==== | ====H260Q==== | ||
<scene name='69/694218/Mutationh260q/1'>Mutation of the histidine component of the catalytic triad, H260, to glutamine</scene> also results in a shift in helix α-9 and <scene name='69/694218/Ag85c-hg/1'>prevents the hydrogen bond formation</scene> between residues His260 and Glu228, thus decreasing enzymatic activity. | <scene name='69/694218/Mutationh260q/1'>Mutation of the histidine component of the catalytic triad, H260, to glutamine</scene> also results in a shift in helix α-9 and <scene name='69/694218/Ag85c-hg/1'>prevents the hydrogen bond formation</scene> between residues His260 and Glu228, thus decreasing enzymatic activity. | ||
| + | |||
| + | ==Student Contributors== | ||
| + | Benjamin Lancaster, Benjamin Zercher, Sarah Zimmerman, & Morgan Blake | ||
</StructureSection> | </StructureSection> | ||
Revision as of 21:12, 15 June 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
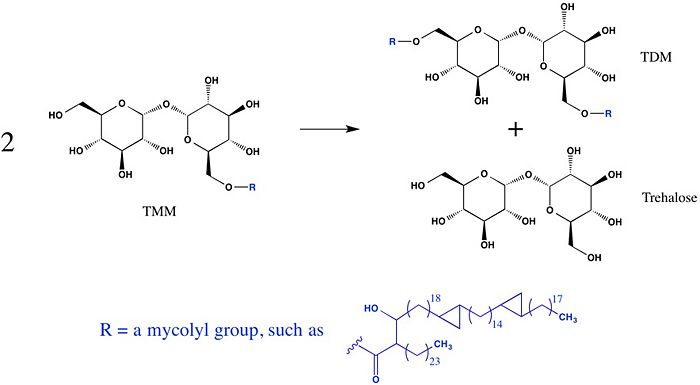
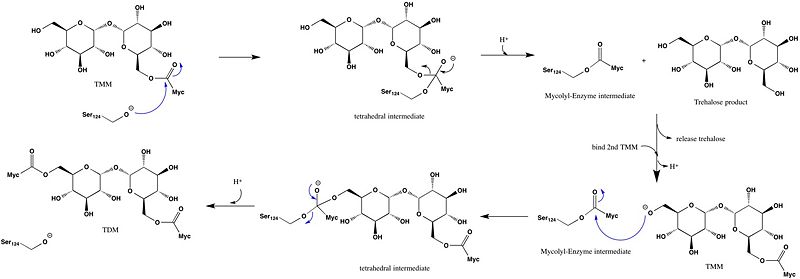
Trehalose-O-mycolyltransferase Ag85C
Introduction
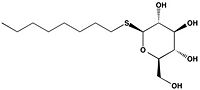
Antigen 85C is one of three homologous protein components of the Ag85 complex in the cell wall of M. tuberculosis. This serine esterase enzyme catalyzes the transfer of mycolyl groups, characteristic components of the cell wall of mycobacteria. Several three dimensional structures of Ag85C have been solved, including the wild type enzyme as well as active site variants due to site-directed mutagenesis and covalent modification.
| |||||||||||
References
- ↑ Jackson M, Raynaud C, Laneelle MA, Guilhot C, Laurent-Winter C, Ensergueix D, Gicquel B, Daffe M. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol Microbiol. 1999 Mar;31(5):1573-87. PMID:10200974
- ↑ Ronning DR, Klabunde T, Besra GS, Vissa VD, Belisle JT, Sacchettini JC. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat Struct Biol. 2000 Feb;7(2):141-6. PMID:10655617 doi:10.1038/72413
- ↑ Ronning DR, Vissa V, Besra GS, Belisle JT, Sacchettini JC. Mycobacterium tuberculosis antigen 85A and 85C structures confirm binding orientation and conserved substrate specificity. J Biol Chem. 2004 Aug 27;279(35):36771-7. Epub 2004 Jun 10. PMID:15192106 doi:http://dx.doi.org/10.1074/jbc.M400811200
- ↑ 4.0 4.1 4.2 4.3 Favrot L, Lajiness DH, Ronning DR. Inactivation of the Mycobacterium tuberculosis Antigen 85 complex by covalent, allosteric inhibitors. J Biol Chem. 2014 Jul 14. pii: jbc.M114.582445. PMID:25028518 doi:http://dx.doi.org/10.1074/jbc.M114.582445