This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Lipase
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
== '''Lipase Catalytic Mechanism''' == | == '''Lipase Catalytic Mechanism''' == | ||
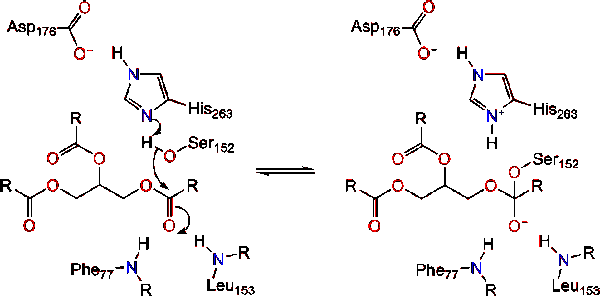
Lipase activation at the lipid-water interface of triacylglycerides, in the presence of colipase and bile salts, is known as interfacial activation. For the hydroloysis reaction to take place, colipase anchors lipase to the lipid-water membrane of the micelle which causes a surface change on lipase. Colipase's four hydrophobic loops interact with the hydrophobic atmosphere of the triacylglyceride. This initiates active site binding to the lipid, and lid opening to reveal a more hydrophobic environment for the triacylglycerol. This in turn, allows the triacylglycerol to interact with key active site residues like the catalytic triad. A diverse array of lipase enzymes can be found in nature. Though the different forms occupy diverse protein scaffolds, most are built upon an alpha/beta hydrolase fold<ref>PMID: 1678899</ref><ref>PMID:1409539 </ref> and possess a [[chymotrypsin]]-like <scene name='Lipase/Catalytic_site_outerview/1'>catalytic triad </scene>comprised of an acidic residue, a histidine, and a serine nucleophile. In the case of horse pancreatic lipase, the catalytic triad is comprised of <scene name='Lipase/Catalytic_triad/4'>Ser 152, Asp 176 and His 263. </scene><ref>PMID:8182745</ref>. This catalytic triad functions like most found in nature. First, aspartic acid forms a hydrogen bond with His 263, increasing the pKa of the histidine imidazole nitrogen. This allows the histidine to act as a powerful general base and deprotonate the serine. The deprotonated serine then can serve as a nucleophile and attack the ester carbonyl of one of the fatty acids on the 1 or 3 carbons of the glycerol backbone of the lipid substrate. Upon attacking the lipid, a negatively charged tetrahedral intermediate is formed (Reaction 1). It is stabilized in the oxyanion hole by two residues: <scene name='Lipase/Catalytic_triad_with_oxyanion/2'>Phe 77 and Leu 153</scene>. | Lipase activation at the lipid-water interface of triacylglycerides, in the presence of colipase and bile salts, is known as interfacial activation. For the hydroloysis reaction to take place, colipase anchors lipase to the lipid-water membrane of the micelle which causes a surface change on lipase. Colipase's four hydrophobic loops interact with the hydrophobic atmosphere of the triacylglyceride. This initiates active site binding to the lipid, and lid opening to reveal a more hydrophobic environment for the triacylglycerol. This in turn, allows the triacylglycerol to interact with key active site residues like the catalytic triad. A diverse array of lipase enzymes can be found in nature. Though the different forms occupy diverse protein scaffolds, most are built upon an alpha/beta hydrolase fold<ref>PMID: 1678899</ref><ref>PMID:1409539 </ref> and possess a [[chymotrypsin]]-like <scene name='Lipase/Catalytic_site_outerview/1'>catalytic triad </scene>comprised of an acidic residue, a histidine, and a serine nucleophile. In the case of horse pancreatic lipase, the catalytic triad is comprised of <scene name='Lipase/Catalytic_triad/4'>Ser 152, Asp 176 and His 263. </scene><ref>PMID:8182745</ref>. This catalytic triad functions like most found in nature. First, aspartic acid forms a hydrogen bond with His 263, increasing the pKa of the histidine imidazole nitrogen. This allows the histidine to act as a powerful general base and deprotonate the serine. The deprotonated serine then can serve as a nucleophile and attack the ester carbonyl of one of the fatty acids on the 1 or 3 carbons of the glycerol backbone of the lipid substrate. Upon attacking the lipid, a negatively charged tetrahedral intermediate is formed (Reaction 1). It is stabilized in the oxyanion hole by two residues: <scene name='Lipase/Catalytic_triad_with_oxyanion/2'>Phe 77 and Leu 153</scene>. | ||
| - | [[Image:M0218. | + | <!-- The next 4 images have been darkened and resized to 600px width. The original files remain with the same filenames minus the "r" at the end ("r" for reduced).--> |
| + | [[Image:M0218.stg01r.gif|center|]] | ||
The carbonyl reforms with the glycerol backbone segment acting as the leaving group (Reaction 2). | The carbonyl reforms with the glycerol backbone segment acting as the leaving group (Reaction 2). | ||
Revision as of 20:58, 21 July 2015
| |||||||||||
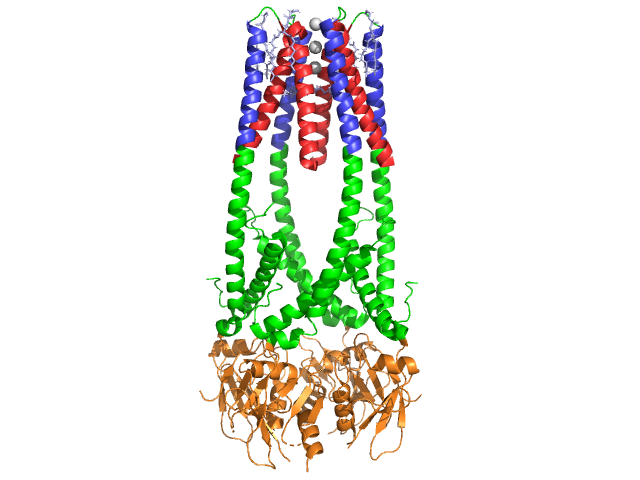
3D Structures of Lipase
Updated on 21-July-2015
References
- ↑ [1] 1HPL PDB SUM
- ↑ [2] A cross-linked complex between horse pancreatic lipase and colipase
- ↑ [3] History of Lipids
- ↑ [4] 1HPL PDB
- ↑ http://www.pdb.org/pdb/explore/explore.do?structureId=1HPL
- ↑ http://www.pdb.org/pdb/explore/remediatedSequence.do?structureId=1HPL
- ↑ http://www.springerlink.com/content/g5h1613440115701/fulltext.pdf
- ↑ Fundamentals of Biochemistry...
- ↑ Thomas, A. etc. "Role of the Lid Hydrophobicity Pattern in Pancreatic Lipase Activity", The Journal of Biological Chemistry, 2005 September 22; 270 (48): 40074-40083.
- ↑ "Colipase". Wikipedia: The Free Encyclopedia. 5 July 2011 [5]
- ↑ "Colipase Residues..."
- ↑ Fundamentals of Biochemistry...
- ↑ Crandall,W., Lowe, M. "Colipase Residues Glu64 and Arg65 Are Essential for Normal Lipase-mediated Fat Digestion in the Presence of Bile Salt Micelles" Journal of Biological Chemistry, 2001, (276) 12505-12512
- ↑ van Tilbeurgh H, etc."Structure of the pancreatic lipase-procolipase complex", 1992 Sep 10;359(6391):159-62. PMID:1522902.[6]
- ↑ http://www.pdb.org/pdb/explore/explore.do?structureId=1ETH
- ↑ http://www.nature.com/nature/journal/v362/n6423/abs/362814a0.html
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al.. The alpha/beta hydrolase fold. Protein Eng. 1992 Apr;5(3):197-211. PMID:1409539
- ↑ Bourne Y, Martinez C, Kerfelec B, Lombardo D, Chapus C, Cambillau C. Horse pancreatic lipase. The crystal structure refined at 2.3 A resolution. J Mol Biol. 1994 May 20;238(5):709-32. PMID:8182745 doi:http://dx.doi.org/10.1006/jmbi.1994.1331
- ↑ [7] 1LPB PDB SUM
- ↑ "Pancreatic lipase". Wikipedia: The Free Encyclopedia. 7 Nov 2011 [8]
- ↑ Kordik, C., Reitz, A. "Pharmacological Treatment of Obesity: Therapeutic Strategies" Journal of Medicinal Chemistry, 1999 (42).
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Quinn R. Murray, Natalie Ziegler, Stephanie Schell, David Canner, Alexander Berchansky, Katelyn Clark, Eric Martz, Leben Tadesse, Joel L. Sussman, Eran Hodis