We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
S100 protein
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='450' side='right' scene='Journal:JBIC:18/Cv/1' caption=''> | |
'''S100 proteins''' are calcium-binding proteins (CBP) found in vertebrates and contain 2 helix-loop-helix calcium binding sites. S100 name is derived from their being 100% soluble in ammonium sulfate. There are at least 21 S100 proteins. Most S100 proteins undergo conformational change upon calcium binding.<br /> | '''S100 proteins''' are calcium-binding proteins (CBP) found in vertebrates and contain 2 helix-loop-helix calcium binding sites. S100 name is derived from their being 100% soluble in ammonium sulfate. There are at least 21 S100 proteins. Most S100 proteins undergo conformational change upon calcium binding.<br /> | ||
| Line 24: | Line 24: | ||
The interacting aromatic residues in <scene name='Journal:JBIC:18/Cv/6'>helix I (Phe-29) and helix IV (Phe-81, Phe-84, Trp-85)</scene> stabilize the interhelical orientation | The interacting aromatic residues in <scene name='Journal:JBIC:18/Cv/6'>helix I (Phe-29) and helix IV (Phe-81, Phe-84, Trp-85)</scene> stabilize the interhelical orientation | ||
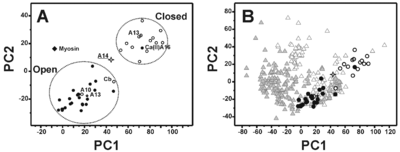
The most remarkable feature of this structural conformation involves the packing of the helices that is reduced with respect to the ‘closed’ structures of the S100 proteins but is still sizably larger than the corresponding ‘open’ structures. At the same time, the analysis of the electrostatic potential surface suggests that the <scene name='Journal:JBIC:18/Cv/2'>S100A14</scene> is <scene name='Journal:JBIC:18/Cv/7'>permanently activated and it is not calcium(II) regulated</scene>. | The most remarkable feature of this structural conformation involves the packing of the helices that is reduced with respect to the ‘closed’ structures of the S100 proteins but is still sizably larger than the corresponding ‘open’ structures. At the same time, the analysis of the electrostatic potential surface suggests that the <scene name='Journal:JBIC:18/Cv/2'>S100A14</scene> is <scene name='Journal:JBIC:18/Cv/7'>permanently activated and it is not calcium(II) regulated</scene>. | ||
| - | + | </StructureSection> | |
== 3D Structures of S100 proteins == | == 3D Structures of S100 proteins == | ||
Revision as of 12:27, 9 August 2015
| |||||||||||
3D Structures of S100 proteins
Updated on 09-August-2015