This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
FK506 binding protein
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
==Wheat FKBP73 and its comparison with human FKBP52<ref >PMID:20306145</ref>== | ==Wheat FKBP73 and its comparison with human FKBP52<ref >PMID:20306145</ref>== | ||
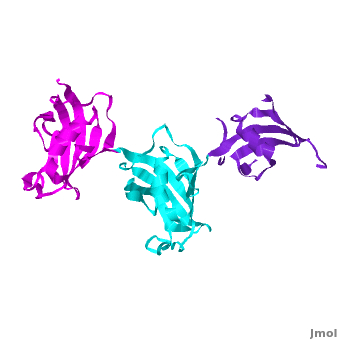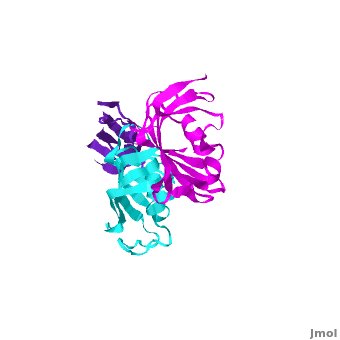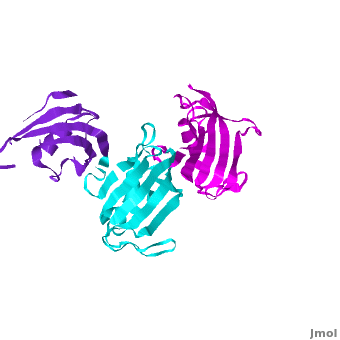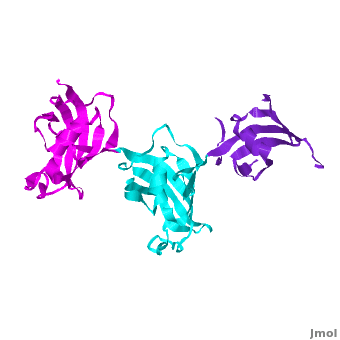
| - | Ribbon representation of the <scene name='3jym/Cv/3'>three FKBP domains</scene>; <font color='blueviolet'><b>wFK73_1 (residues 1–148) in blueviolet</b></font>, < | + | Ribbon representation of the <scene name='3jym/Cv/3'>three FKBP domains</scene>; <font color='blueviolet'><b>wFK73_1 (residues 1–148) in blueviolet</b></font>, <span style="color:cyan;background-color:black;font-weight:bold;">wFK73_2 (residues 149–266) in cyan</span> and <font color='magenta'><b>wFK73_3 (residues 267–386) in magenta</b></font> ([[3jym]]). The wFK73_1 domain exhibits electron density only between residues 33–38, 54–69 and 87–148. The bulges and the flaps as well as the N- and C-termini are labeled. The three FK506 binding (FK) domains of wFKBP73 are held together mainly by <scene name='3jym/Cv/4'>salt bridge networks</scene> situated between each pair of domains. The wFK73_2-wFK73_1 domains are held by a salt bridge between Lys162–Glu62, and a salt bridge network between Arg151–Asp61 and Glu58. The interface between wFK73_2-wFK73_3 is held by two salt bridges between Lys204–Glu269, and Glu178–Lys279. The interactions Lys162–Glu62 and Glu178–Lys279, involve conserved residues (Glu62 from wFK73_1 and Glu178 from wFK73_2, Lys162 from wFK73_2 and Lys279 from wFK73_3). |
{{Clear}} | {{Clear}} | ||
| - | The 3D structures of several FKBP family members from various species are solved, most of them comprise 1-2 FK domains (''e.g.'' human FKBP52, known also as FKBP4), while wFKBP73 has 3 FK domains which is characteristic to plants. A sequence-based structure comparison between each of the 3 FK domains of wFKBP73 and the 2 FK domains of hFKBP52 ([[1q1c]]) was performed. All 3 FK domains of wFKBP73 adopt a typical FK fold exhibiting significant diversity when superimposed. They are arranged in a linear manner in space as observed in the 2 FK domains of hFKBP52. While the 2 FK domains of hFKBP52 are in the same orientation, the orientation between any 2 consecutive wFK73 domains is different than that between the two FK domains of hFKBP52. <scene name='3jym/Al/2'>Superposition</scene> of the <font color='blueviolet'><b>wFK73_1 (in blueviolet)</b></font> and < | + | The 3D structures of several FKBP family members from various species are solved, most of them comprise 1-2 FK domains (''e.g.'' human FKBP52, known also as FKBP4), while wFKBP73 has 3 FK domains which is characteristic to plants. A sequence-based structure comparison between each of the 3 FK domains of wFKBP73 and the 2 FK domains of hFKBP52 ([[1q1c]]) was performed. All 3 FK domains of wFKBP73 adopt a typical FK fold exhibiting significant diversity when superimposed. They are arranged in a linear manner in space as observed in the 2 FK domains of hFKBP52. While the 2 FK domains of hFKBP52 are in the same orientation, the orientation between any 2 consecutive wFK73 domains is different than that between the two FK domains of hFKBP52. <scene name='3jym/Al/2'>Superposition</scene> of the <font color='blueviolet'><b>wFK73_1 (in blueviolet)</b></font> and <span style="color:cyan;background-color:black;font-weight:bold;">wFK73_2 (in cyan)</span> domains on hFK52_1 (in yellow) and <font color='blue'><b>hFK52_2 (in blue)</b></font> revealed that while <span style="color:cyan;background-color:black;font-weight:bold;">wFK73_2</span> is perfectly aligned with <font color='blue'><b>hFK52_2</b></font>, N-terminal <font color='blueviolet'><b>wFK73_1</b></font> does not align with hFK52_1 (yellow). Similarly, <scene name='3jym/Al/3'>superposition</scene> of the <span style="color:cyan;background-color:black;font-weight:bold;">wFK73_2</b></span> and <font color='magenta'><b>wFK73_3 (in magenta)</b></font> domains on hFKBP52 revealed that while <span style="color:cyan;background-color:black;font-weight:bold;">wFK73_2</span> is perfectly aligned with hFK52_1 (in yellow), <font color='magenta'><b>wFK73_3</b></font> does not align with <font color='blue'><b>hFK52_2</b></font>. This unique arrangement of wFKBP73 causes that the α-helices of <scene name='3jym/Al/4'>wFKBP73 3 FK domains</scene> are exposed on the same surface, while the <scene name='3jym/Al/5'>2 α-helices of hFK52</scene> are presented on opposite surfaces. |
{{Clear}} | {{Clear}} | ||
| - | It was shown that 12 conserved residues (for wFK73_1 domain they are Tyr67, Phe77, Asp78, Arg83, Phe87, Gln95, Val96, Ile97, Trp100, Tyr123, Ile132, and Phe140) of the FK1 domains of hFKBP12, 13, 25, 51 and 52, are involved in binding the FK506 or rapamycin. Since only the FK1 domains contain all the conserved amino acids (in contrast to FK2 and/or FK3 domains), only they exhibit PPIase activity, which can be inhibited by the binding of the drugs FK506, and rapamycin. These conserved residues form the hydrophobic cavity. The structure of hFKBP12 ([[2ppn]]) demonstrates a good example of this <scene name='3jym/Cavity/1'>cavity</scene>. All these residues are conserved in the wFK73_1 domain, it could be assumed that a similar cavity is also formed in wFK73_1, although some of these residues are missing electron density in the wFK73 structure and, therefore, it can not be seen. Domain <font color='magenta'><b>wFK73_3</b></font> has <scene name='3jym/Cavity/3'>narrower cavity</scene>, whereas < | + | It was shown that 12 conserved residues (for wFK73_1 domain they are Tyr67, Phe77, Asp78, Arg83, Phe87, Gln95, Val96, Ile97, Trp100, Tyr123, Ile132, and Phe140) of the FK1 domains of hFKBP12, 13, 25, 51 and 52, are involved in binding the FK506 or rapamycin. Since only the FK1 domains contain all the conserved amino acids (in contrast to FK2 and/or FK3 domains), only they exhibit PPIase activity, which can be inhibited by the binding of the drugs FK506, and rapamycin. These conserved residues form the hydrophobic cavity. The structure of hFKBP12 ([[2ppn]]) demonstrates a good example of this <scene name='3jym/Cavity/1'>cavity</scene>. All these residues are conserved in the wFK73_1 domain, it could be assumed that a similar cavity is also formed in wFK73_1, although some of these residues are missing electron density in the wFK73 structure and, therefore, it can not be seen. Domain <font color='magenta'><b>wFK73_3</b></font> has <scene name='3jym/Cavity/3'>narrower cavity</scene>, whereas <span style="color:cyan;background-color:black;font-weight:bold;">wFK73_2</span> <scene name='3jym/Cavity/4'>lacks this cavity at all</scene>. Conserved residues are colored yellow. So, the lack of drug binding of the wFK73_2 and wFK73_3 domains could be explained by the absence of the conserved drug binding residues. This is in agreement with the fact that the FK2 domains of hFKBP51 and hFKBP52 and the single FK domains of FKBP38, DmFKBP45 and AtFKBP42, all lacking the conserved residues, do not exhibit drug binding. |
==SlyD<ref >DOI 10.1007/s00775-011-0855-y</ref>== | ==SlyD<ref >DOI 10.1007/s00775-011-0855-y</ref>== | ||
Revision as of 12:39, 9 August 2015
| |||||||||||
3D Structures of FKBP
Updated on 09-August-2015