Inosine monophosphate dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Structural highlights == | == Structural highlights == | ||
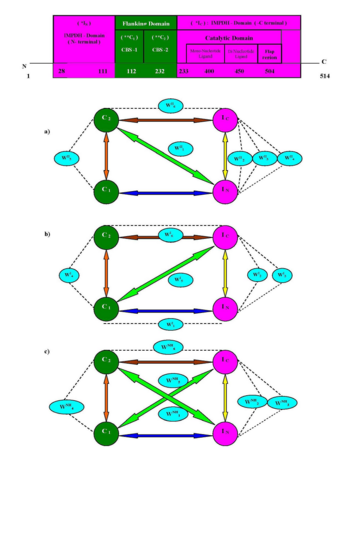
| - | === An Insight to the Dynamics of Conserved Water Mediated Salt Bridge Interaction and Inter-Domain Recognition in hIMPDH Isoforms | + | === An Insight to the Dynamics of Conserved Water Mediated Salt Bridge Interaction and Inter-Domain Recognition in hIMPDH Isoforms <ref>DOI 10.1080/07391102.2012.712458</ref>=== |
| - | + | ||
| - | + | ||
| - | + | ||
Dynamic personality of protein is inherent in Nature and necessary for performing the chemical reaction in living cells. The functions of proteins / enzymes and their physical properties are also governed by their dynamic characters which requires addition of a fourth dimension, time and atomic resolution. Detail investigation of dynamic and X-ray structures of protein or their complexes with different molecules can unfold the structure-function relationship of protein. Presumably, the dynamic propensity of protein is not only determine the chemical transformations/ reaction mechanism of biosynthetic processes but it also vividly illustrated the insight of topological feature of inhibitor. It is interesting in enzymology that Nature also installs some similar proteins which are almost have identical function, catalytic machinery, 3D-structures and are known as isoform. The salt bridge interaction (acidic and basic residues) as well as their conjugation through amphoteric water molecules (Acid---Water---Base) may be indispensable component of protein recognition, domain assemble and structure stabilization. The differential inter domain recognition through conserved water mediated salt bridge interaction in type -I and II isoforms (of human IMPDH) may open a new avenue towards their inhibitor development. | Dynamic personality of protein is inherent in Nature and necessary for performing the chemical reaction in living cells. The functions of proteins / enzymes and their physical properties are also governed by their dynamic characters which requires addition of a fourth dimension, time and atomic resolution. Detail investigation of dynamic and X-ray structures of protein or their complexes with different molecules can unfold the structure-function relationship of protein. Presumably, the dynamic propensity of protein is not only determine the chemical transformations/ reaction mechanism of biosynthetic processes but it also vividly illustrated the insight of topological feature of inhibitor. It is interesting in enzymology that Nature also installs some similar proteins which are almost have identical function, catalytic machinery, 3D-structures and are known as isoform. The salt bridge interaction (acidic and basic residues) as well as their conjugation through amphoteric water molecules (Acid---Water---Base) may be indispensable component of protein recognition, domain assemble and structure stabilization. The differential inter domain recognition through conserved water mediated salt bridge interaction in type -I and II isoforms (of human IMPDH) may open a new avenue towards their inhibitor development. | ||
Revision as of 12:09, 10 August 2015
| |||||||||||
3D structures of inosine monophosphate dehydrogenase
Updated on 10-August-2015
References
- ↑ Bairagya HR, Mukhopadhyay BP. An insight to the dynamics of conserved water-mediated salt bridge interaction and interdomain recognition in hIMPDH isoforms. J Biomol Struct Dyn. 2012 Aug 28. PMID:22928911 doi:10.1080/07391102.2012.712458