Ornithine decarboxylase
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
===CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION ([[7odc]]) <ref>PMID:10378276</ref><ref>PMID:18369191</ref>=== | ===CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION ([[7odc]]) <ref>PMID:10378276</ref><ref>PMID:18369191</ref>=== | ||
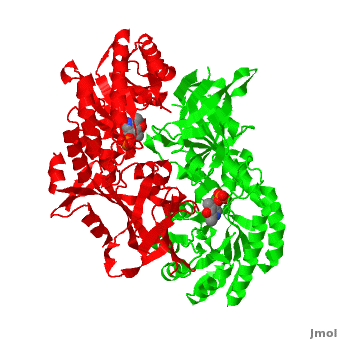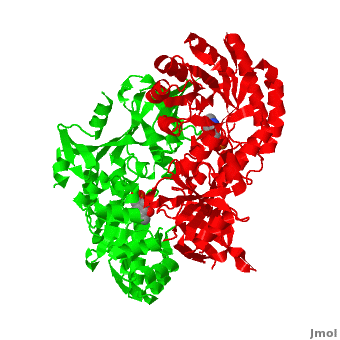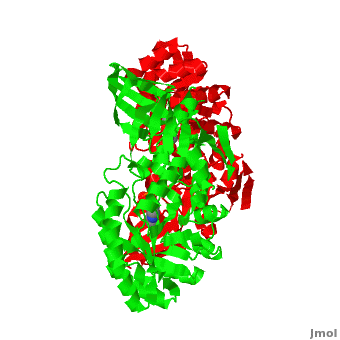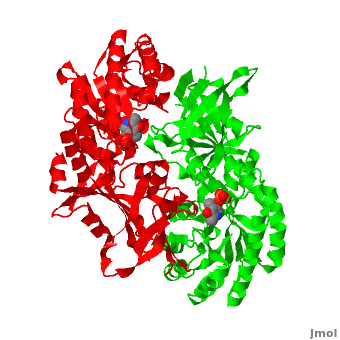
| - | Although, [[7odc]] is a 1 chain structure, the biological relevant molecule for [[7odc]] can be assembled from the contents of the deposited coordinates by the application of crystallographic symmetry operations to give a dimer. It can be [http://www.ebi.ac.uk/pdbe/pqs/macmol/7odc.mmol downloaded]. A sequence alignment and structural comparison of mouse [http://en.wikipedia.org/wiki/Ornithine_decarboxylase ornithine decarboxylase] (mODC) to mouse [[Antizyme Inhibitor]] (AzI, [[3btn]]) show high sequence identity (~50%) and structural similarity between mODC and AzI monomers (RMSD value is 1.6 Å). The <scene name='Antizyme_Inhibitor/Azi_odc/10'>structural comparison</scene> of mODC (<font color='red'><b>red</b></font> and <font color='lime'><b>lime</b></font>) to mouse AzI crystallographic dimer ( | + | Although, [[7odc]] is a 1 chain structure, the biological relevant molecule for [[7odc]] can be assembled from the contents of the deposited coordinates by the application of crystallographic symmetry operations to give a dimer. It can be [http://www.ebi.ac.uk/pdbe/pqs/macmol/7odc.mmol downloaded]. A sequence alignment and structural comparison of mouse [http://en.wikipedia.org/wiki/Ornithine_decarboxylase ornithine decarboxylase] (mODC) to mouse [[Antizyme Inhibitor]] (AzI, [[3btn]]) show high sequence identity (~50%) and structural similarity between mODC and AzI monomers (RMSD value is 1.6 Å). The <scene name='Antizyme_Inhibitor/Azi_odc/10'>structural comparison</scene> of mODC (<font color='red'><b>red</b></font> and <font color='lime'><b>lime</b></font>) to mouse AzI crystallographic dimer (<span style="color:cyan;background-color:black;font-weight:bold;">mAzI, cyan</span> and <font color='blueviolet'><b>blueviolet</b></font>) is shown. Superposition of the <scene name='Antizyme_Inhibitor/Azi_odc/11'>interface</scene> of mAzI and mODC showing the inter-subunit variable loops (AzI residues 355–362 and 387–401). <font color='black'><b>AzI loops</b></font> are in <font color='black'><b>black</b></font>, and <font color='black'><b>ODC loops</b></font> are in <font color='black'><b>yellow</b></font>. |
| - | The two AzI monomers (< | + | The two AzI monomers (<span style="color:cyan;background-color:black;font-weight:bold;">cyan</span>, <font color='blueviolet'><b>blueviolet</b></font>) have only <scene name='Antizyme_Inhibitor/Azi_odc1/1'>43 contacting residues</scene> (< 3.5 Å apart), while there are more contacts (83) between the two monomers of <scene name='Antizyme_Inhibitor/Azi_odc1/2'>mODC</scene> (<font color='red'><b>red</b></font>, <font color='lime'><b>lime</b></font>). Moreover, the surface area buried by the two mODC monomers is significantly larger than the one buried by the AzI monomers. These features explain a very weak crystallographic AzI dimer. |
The zipper, formed by conserved [http://en.wikipedia.org/wiki/Hydrophobe hydrophobic] residues in mODC, stabilizes its dimeric structure. These residues involve F397(B), Y323(B), Y331(A), Y331(B), Y323(A), and F397(A) (the names of the chains are in brackets). The residue Y331 in the <scene name='Antizyme_Inhibitor/Azi_odc1/3'>ODC zipper</scene> is substituted by S329 in AzI and interferes with the formation of a similar zipper in AzI. Hence, in <scene name='Antizyme_Inhibitor/Azi_odc1/4'>mAzI</scene> this hydrophobic zipper is absent. Many residues, participating in the ODC interdimer interface interactions, are conserved among the ODCs from variuos organisms, but in AzI these residues are not conserved. Furthermore, the AzI conserved residues do not participate in interdimer interactions. For example, mODC possesses <scene name='Antizyme_Inhibitor/Azi_odc1/5'>two salt bridges</scene> (K169–D364 and D134–K294) stabilizing the ODC [http://en.wikipedia.org/wiki/Dimer homodimer]. In AzI, these 4 corresponding residues (<scene name='Antizyme_Inhibitor/Azi_odc1/6'>K169-D362 and D134-K291</scene>, respectively) are also present, but are too far apart to form a salt bridge. The two AzI monomers are positioned farther apart, in comparison ot ODC monomers, preventing the formation of interdimer interactions. | The zipper, formed by conserved [http://en.wikipedia.org/wiki/Hydrophobe hydrophobic] residues in mODC, stabilizes its dimeric structure. These residues involve F397(B), Y323(B), Y331(A), Y331(B), Y323(A), and F397(A) (the names of the chains are in brackets). The residue Y331 in the <scene name='Antizyme_Inhibitor/Azi_odc1/3'>ODC zipper</scene> is substituted by S329 in AzI and interferes with the formation of a similar zipper in AzI. Hence, in <scene name='Antizyme_Inhibitor/Azi_odc1/4'>mAzI</scene> this hydrophobic zipper is absent. Many residues, participating in the ODC interdimer interface interactions, are conserved among the ODCs from variuos organisms, but in AzI these residues are not conserved. Furthermore, the AzI conserved residues do not participate in interdimer interactions. For example, mODC possesses <scene name='Antizyme_Inhibitor/Azi_odc1/5'>two salt bridges</scene> (K169–D364 and D134–K294) stabilizing the ODC [http://en.wikipedia.org/wiki/Dimer homodimer]. In AzI, these 4 corresponding residues (<scene name='Antizyme_Inhibitor/Azi_odc1/6'>K169-D362 and D134-K291</scene>, respectively) are also present, but are too far apart to form a salt bridge. The two AzI monomers are positioned farther apart, in comparison ot ODC monomers, preventing the formation of interdimer interactions. | ||
Revision as of 12:17, 10 August 2015
| |||||||||||
3D structures of ornithine decarboxylase
Updated on 10-August-2015
References
- ↑ Kern AD, Oliveira MA, Coffino P, Hackert ML. Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure. 1999 May;7(5):567-81. PMID:10378276
- ↑ Albeck S, Dym O, Unger T, Snapir Z, Bercovich Z, Kahana C. Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci. 2008 May;17(5):793-802. Epub 2008 Mar 27. PMID:18369191 doi:10.1110/ps.073427208
- ↑ Sanchita, Chauhan R, Soni G, Sudhamalla B, Sharma A. Docking and molecular dynamics studies of peptide inhibitors of ornithine decarboxylase: a rate-limiting enzyme for the metabolism of Fusarium solani. J Biomol Struct Dyn. 2012 Sep 13. PMID:22970930 doi:10.1080/07391102.2012.718526