Hen Egg-White (HEW) Lysozyme
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
All proteins consist of carbon, hydrogen, nitrogen, oxygen, and sulfur, as do most organic molecules. Enzymes are composed in such a way as to maximize their reactivity with their desired substrate, increasing the efficiency of biological reactions. The <scene name='Sandbox_39/Elements/1'>composition of lysozyme</scene> can be seen on the left, with the carbon atoms outlined in gray, oxygen atoms in red, nitrogen atoms in blue, sulfur atoms in yellow, and the three-letter abbreviation for the <scene name='Sandbox_39/Amino_acid_residues/1'>amino acid residues</scene> in purple. Lysozyme, like all proteins, also contains <scene name='Sandbox_39/C_and_n_terminal_residues/1'> C and N termini </scene>, and these can be seen by following the colors of the rainbow across the molecule. Starting at the red end, the C terminal end, one can work the entire way through to the N terminal end, showing the folding pattern and chain of the protein. | All proteins consist of carbon, hydrogen, nitrogen, oxygen, and sulfur, as do most organic molecules. Enzymes are composed in such a way as to maximize their reactivity with their desired substrate, increasing the efficiency of biological reactions. The <scene name='Sandbox_39/Elements/1'>composition of lysozyme</scene> can be seen on the left, with the carbon atoms outlined in gray, oxygen atoms in red, nitrogen atoms in blue, sulfur atoms in yellow, and the three-letter abbreviation for the <scene name='Sandbox_39/Amino_acid_residues/1'>amino acid residues</scene> in purple. Lysozyme, like all proteins, also contains <scene name='Sandbox_39/C_and_n_terminal_residues/1'> C and N termini </scene>, and these can be seen by following the colors of the rainbow across the molecule. Starting at the red end, the C terminal end, one can work the entire way through to the N terminal end, showing the folding pattern and chain of the protein. | ||
| - | Lysozyme contains five <scene name='Sandbox_38/A/2'>alpha helical</scene> regions and five regions containing <scene name='Sandbox_38/B/1'>beta sheets</scene> as displayed in this <scene name='Sandbox_38/Alphab/1'>image</scene>. Linking these secondary structures, a number of beta turns and a large number of random coils make up the remainder of the polypeptide backbone. The polypeptide backbone of lysozyme involved in the 3 antiparallel beta sheets display the beta hairpin motif of supersecondary structure. This depiction of lysozyme contains an antiparallel beta-pleated sheet, which contributes greatly to the stability of the molecule by providing the correct alignment of hydrogen bonds. Lysozyme also contains a great deal of random coil, which is seen in the white regions of the molecule. | + | Lysozyme contains five <scene name='Sandbox_38/A/2'>alpha helical</scene> regions and five regions containing <scene name='Sandbox_38/B/1'>beta sheets</scene> as displayed in this <scene name='Sandbox_38/Alphab/1'>image</scene>. |
| + | |||
| + | Linking these secondary structures, a number of beta turns and a large number of random coils make up the remainder of the polypeptide backbone. The polypeptide backbone of lysozyme involved in the 3 antiparallel beta sheets display the beta hairpin motif of supersecondary structure. This depiction of lysozyme contains an antiparallel beta-pleated sheet, which contributes greatly to the stability of the molecule by providing the correct alignment of hydrogen bonds. Lysozyme also contains a great deal of random coil, which is seen in the white regions of the molecule. | ||
=== Intermolecular Interactions === | === Intermolecular Interactions === | ||
| Line 47: | Line 49: | ||
[[Image:jrip.jpg|thumb|left|250px|Mechanism of Lysozyme<ref>Image from: http://www.google.com/imgres?imgurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/pics-and-strucs/lysozyme-mech.gif&imgrefurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm&usg=__ormapG4XKg-tR5GrMSOdSMTV4vE=&h=603&w=801&sz=7&hl=en&start=17&zoom=1&tbnid=nvr9gvFrUILDkM:&tbnh=143&tbnw=189&prev=/images%3Fq%3DThe%2Blysozyme%2Breaction%2Bmechanism%26um%3D1%26hl%3Den%26sa%3DN%26biw%3D1280%26bih%3D647%26tbs%3Disch:10%2C304&um=1&itbs=1&iact=hc&vpx=521&vpy=349&dur=448&hovh=191&hovw=254&tx=140&ty=48&ei=JQ_LTPKzLIjCsAPkzt2KDg&oei=IA_LTP74OsG78gapm-GFAQ&esq=2&page=2&ndsp=18&ved=1t:429,r:2,s:17&biw=1280&bih=647</ref>]] | [[Image:jrip.jpg|thumb|left|250px|Mechanism of Lysozyme<ref>Image from: http://www.google.com/imgres?imgurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/pics-and-strucs/lysozyme-mech.gif&imgrefurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm&usg=__ormapG4XKg-tR5GrMSOdSMTV4vE=&h=603&w=801&sz=7&hl=en&start=17&zoom=1&tbnid=nvr9gvFrUILDkM:&tbnh=143&tbnw=189&prev=/images%3Fq%3DThe%2Blysozyme%2Breaction%2Bmechanism%26um%3D1%26hl%3Den%26sa%3DN%26biw%3D1280%26bih%3D647%26tbs%3Disch:10%2C304&um=1&itbs=1&iact=hc&vpx=521&vpy=349&dur=448&hovh=191&hovw=254&tx=140&ty=48&ei=JQ_LTPKzLIjCsAPkzt2KDg&oei=IA_LTP74OsG78gapm-GFAQ&esq=2&page=2&ndsp=18&ved=1t:429,r:2,s:17&biw=1280&bih=647</ref>]] | ||
| - | + | {{Clear}} | |
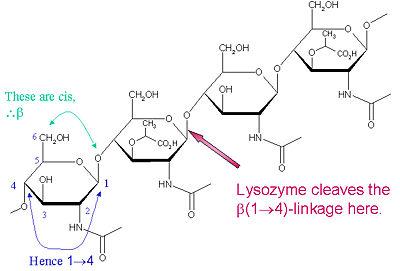
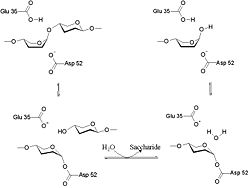
The lysozyme mechanism of action results in the hydrolysis of a glycoside (hence the familial distinction of lysozyme as a glycosylase<ref>Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/</ref>), which corresponds to the conversion of an acetal to a hemiacetal, which reaction (general degradation of glycosidic bond to units "capped" by newly formed hydroxyl groups) necessitates acid catalysis, since the conversion of acetal to hemiacetal involves the protonation of the reactant oxygen prior to actual bond cleavage. <ref>Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.</ref>. Furthermore, the transition state obtained from this protonation is a covalent, oxonium ion, intermediate that must obtain resonance stabilization. The need for some means of acid catalysis and covalent resonance stabilization is adequately provided by the Glu 35 and Asp 52 residues of lysozyme, respectively. The reaction mechanism of lysozyme is demonstrated on the left. The reaction begins at the upper left-hand side, and proceeds according to reaction arrows. Lysozyme works by hydrolyzing the glycosidic bond, distorting the bond between the NAM and NAG. This produces a glycosyl enzyme intermediate, which reacts with a water molecule to produce the product and the unchanged enzyme. The <scene name='Hen_Egg-White_(HEW)_Lysozyme/Act_site/2'>active site</scene> of lysozyme is formulated as a prominent cleft outlined by the two aforementioned catalytic amino acids, Glu 35 and Asp 52. The active site is geometrically bent to augment ligand binding, and the two amino acids interact with the ligand in the binding site. Asp52 is depicted in red, and Glu35 is depicted in green. | The lysozyme mechanism of action results in the hydrolysis of a glycoside (hence the familial distinction of lysozyme as a glycosylase<ref>Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/</ref>), which corresponds to the conversion of an acetal to a hemiacetal, which reaction (general degradation of glycosidic bond to units "capped" by newly formed hydroxyl groups) necessitates acid catalysis, since the conversion of acetal to hemiacetal involves the protonation of the reactant oxygen prior to actual bond cleavage. <ref>Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.</ref>. Furthermore, the transition state obtained from this protonation is a covalent, oxonium ion, intermediate that must obtain resonance stabilization. The need for some means of acid catalysis and covalent resonance stabilization is adequately provided by the Glu 35 and Asp 52 residues of lysozyme, respectively. The reaction mechanism of lysozyme is demonstrated on the left. The reaction begins at the upper left-hand side, and proceeds according to reaction arrows. Lysozyme works by hydrolyzing the glycosidic bond, distorting the bond between the NAM and NAG. This produces a glycosyl enzyme intermediate, which reacts with a water molecule to produce the product and the unchanged enzyme. The <scene name='Hen_Egg-White_(HEW)_Lysozyme/Act_site/2'>active site</scene> of lysozyme is formulated as a prominent cleft outlined by the two aforementioned catalytic amino acids, Glu 35 and Asp 52. The active site is geometrically bent to augment ligand binding, and the two amino acids interact with the ligand in the binding site. Asp52 is depicted in red, and Glu35 is depicted in green. | ||
Revision as of 11:14, 3 September 2015
| |||||||||||
See Also
References
- ↑ Lysozyme. 2010. Citizendium.org. http://en.citizendium.org/wiki/Lysozyme
- ↑ Lysozyme. 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Bugg, T. 1997. An Introduction to Enzyme and Coenzyme Chemistry. Blackwell Science Ltd., Oxford
- ↑ 1967. Proc R Soc Lond B Bio 167 (1009): 389–401.
- ↑ Image from: http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm
- ↑ http://mcdb-webarchive.mcdb.ucsb.edu/sears/biochemistry/tw-enz/lysozyme/HEWL/lysozyme-overview.htm
- ↑ http://www.worthington-biochem.com/ly/default.html
- ↑ Image from: http://www.google.com/imgres?imgurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/pics-and-strucs/lysozyme-mech.gif&imgrefurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm&usg=__ormapG4XKg-tR5GrMSOdSMTV4vE=&h=603&w=801&sz=7&hl=en&start=17&zoom=1&tbnid=nvr9gvFrUILDkM:&tbnh=143&tbnw=189&prev=/images%3Fq%3DThe%2Blysozyme%2Breaction%2Bmechanism%26um%3D1%26hl%3Den%26sa%3DN%26biw%3D1280%26bih%3D647%26tbs%3Disch:10%2C304&um=1&itbs=1&iact=hc&vpx=521&vpy=349&dur=448&hovh=191&hovw=254&tx=140&ty=48&ei=JQ_LTPKzLIjCsAPkzt2KDg&oei=IA_LTP74OsG78gapm-GFAQ&esq=2&page=2&ndsp=18&ved=1t:429,r:2,s:17&biw=1280&bih=647
- ↑ Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Johnson LN, Phillips DC. Structure of some crystalline lysozyme-inhibitor complexes determined by X-ray analysis at 6 Angstrom resolution. Nature. 1965 May 22;206(986):761-3. PMID:5840126
- ↑ Phillips, D. C. The hen egg white lysozyme molecule. Proc. Natl Acad. Sci. USA 57, 483-495 (1967)
- ↑ coordinates of the model kindly provided by Louise Johnson
- ↑ Vocadlo DJ, Davies GJ, Laine R, Withers SG. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature. 2001 Aug 23;412(6849):835-8. PMID:11518970 doi:10.1038/35090602
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Michal Harel, Anne Goodling, Alexander Berchansky, Eric Martz, Joel L. Sussman, Karl Oberholser, Jaime Prilusky, John Ripollone, Daniel Kreider, Judy Voet, John S. de Banzie