Potassium Channel
From Proteopedia
| Line 1: | Line 1: | ||
<StructureSection load='' size='500' side='right' caption='Structure of the Potassium Channel complex with K+ ions, ([[2r9r]])' scene='Potassium_Channel/Opening/1'> | <StructureSection load='' size='500' side='right' caption='Structure of the Potassium Channel complex with K+ ions, ([[2r9r]])' scene='Potassium_Channel/Opening/1'> | ||
| - | [[Image:2r9r Picture Proteopedia2.png|250px|left]] [[Potassium Channel]]'''s''' control cell membrane electric potentials by selectively allowing diffusion of K<sup>+</sup> across the membrane.<ref name="Zhou">PMID: 11689936</ref> K<sup>+</sup> Channels extend across the cell membrane, a 40Å thick lipid bilayer which ions cannot cross.<ref name="Doyle">PMID: 9525859</ref> Potassium homeostasis is crucial for nearly all living cells, but is particularly important for the correct function of neurons. Neurons produce electrical impulses known as action potentials, allow for cellular communication processes like neurotransmitter release or to initiate intercellular processes muscle contraction. At the onset of an action potential, sodium ions flood across the plasma membrane of neurons via sodium channels. The change in polarity of the plasma membrane caused by the sodium ion influx inactivates sodium channels. Potassium channels subsequently open allowing the selective diffusion of K<sup>+</sup> ions across the plasma membrane, returning the membrane polarity to neutral. After the action potential has passed, channels recreate the high potassium concentration within the cell in preparation for the next stiumulus.<ref>PMID:12721618</ref> Mutations in voltage-gated potassium channel KCNC3 have been linked with [[Neurodevelopmental Disorders|neurodevelopmental disorders]] and neurodegeneration.<ref>PMID: 16501573</ref> | + | [[Image:2r9r Picture Proteopedia2.png|250px|left]] |
| + | {{Clear}} | ||
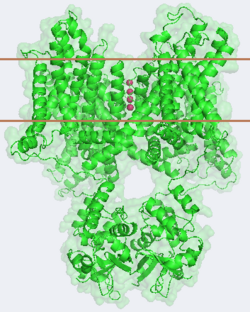
| + | [[Potassium Channel]]'''s''' control cell membrane electric potentials by selectively allowing diffusion of K<sup>+</sup> across the membrane.<ref name="Zhou">PMID: 11689936</ref> K<sup>+</sup> Channels extend across the cell membrane, a 40Å thick lipid bilayer which ions cannot cross.<ref name="Doyle">PMID: 9525859</ref> Potassium homeostasis is crucial for nearly all living cells, but is particularly important for the correct function of neurons. Neurons produce electrical impulses known as action potentials, allow for cellular communication processes like neurotransmitter release or to initiate intercellular processes muscle contraction. At the onset of an action potential, sodium ions flood across the plasma membrane of neurons via sodium channels. The change in polarity of the plasma membrane caused by the sodium ion influx inactivates sodium channels. Potassium channels subsequently open allowing the selective diffusion of K<sup>+</sup> ions across the plasma membrane, returning the membrane polarity to neutral. After the action potential has passed, channels recreate the high potassium concentration within the cell in preparation for the next stiumulus.<ref>PMID:12721618</ref> Mutations in voltage-gated potassium channel KCNC3 have been linked with [[Neurodevelopmental Disorders|neurodevelopmental disorders]] and neurodegeneration.<ref>PMID: 16501573</ref> | ||
Potassium channels possess two traits that are seemingly mutually exclusive. Firstly, potassium channels have exquisite selectivity, with an amazing 10,000 fold selectivity for K<sup>+</sup> ions over sodium ions. Considering the only difference by which potassium ions can be differentiated from sodium ions is potassium ions’ 1.33Å Pauling radius vs. Sodium’s .95Å radius, the selectivity of potassium channels is remarkable.<ref name="Doyle"/> Second, despite its remarkable selectivity, potassium channels allow for the transfer of K<sup>+</sup> ions across the cell membrane at a rate of nearly 10<sup>8</sup> per second, nearly at the diffusion rate limit.<ref name="Long">PMID: 18004376</ref> Potassium channels are able to achieve these remarkable feats due to its amazing structural architecture, which contains several features which not only can sense the voltage potential across a membrane, but also selectively ferry K<sup>+</sup> ions without any outside energy expenditure. | Potassium channels possess two traits that are seemingly mutually exclusive. Firstly, potassium channels have exquisite selectivity, with an amazing 10,000 fold selectivity for K<sup>+</sup> ions over sodium ions. Considering the only difference by which potassium ions can be differentiated from sodium ions is potassium ions’ 1.33Å Pauling radius vs. Sodium’s .95Å radius, the selectivity of potassium channels is remarkable.<ref name="Doyle"/> Second, despite its remarkable selectivity, potassium channels allow for the transfer of K<sup>+</sup> ions across the cell membrane at a rate of nearly 10<sup>8</sup> per second, nearly at the diffusion rate limit.<ref name="Long">PMID: 18004376</ref> Potassium channels are able to achieve these remarkable feats due to its amazing structural architecture, which contains several features which not only can sense the voltage potential across a membrane, but also selectively ferry K<sup>+</sup> ions without any outside energy expenditure. | ||
| Line 16: | Line 18: | ||
Overall, Potassium channels are remarkable structures that allow for near diffusion limit transfer of molecules with sub-angstrom specificity. Our understanding of the structure of Potassium Channels has opened up the potential for [[Pharmaceutical drugs|therapeutic intervention]] into Potassium channel related diseases. | Overall, Potassium channels are remarkable structures that allow for near diffusion limit transfer of molecules with sub-angstrom specificity. Our understanding of the structure of Potassium Channels has opened up the potential for [[Pharmaceutical drugs|therapeutic intervention]] into Potassium channel related diseases. | ||
</StructureSection> | </StructureSection> | ||
| - | + | __NOTOC__ | |
==Page Development== | ==Page Development== | ||
This article was developed based on lectures given in Chemistry 543 by Prof. Clarence E. Schutt at Princeton University. | This article was developed based on lectures given in Chemistry 543 by Prof. Clarence E. Schutt at Princeton University. | ||
Revision as of 12:30, 3 September 2015
| |||||||||||
Page Development
This article was developed based on lectures given in Chemistry 543 by Prof. Clarence E. Schutt at Princeton University.
Potassium channels (KCh) are subdivided into voltage-gated KCh and calcium-dependent KCh. The latter are subdivided into high- (BK, LKCa), intermediate- and small-conductance KCh (human SK1, rat SK2, SKCa). The T1 domain is a highly conserved N-terminal domain which is responsible for driving the tetramerization of the KCh α subunit. The inward rectifier KCh (IRK) passes current more easily in the inward direction. MthK is a calcium-dependent KCh from Methanobacterium thermoautrophicum.
3D structures of Potassium Channels
Updated on 03-September-2015
Additional Resources
References
- ↑ 1.0 1.1 Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001 Nov 1;414(6859):43-8. PMID:11689936 doi:http://dx.doi.org/10.1038/35102009
- ↑ 2.0 2.1 2.2 2.3 Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998 Apr 3;280(5360):69-77. PMID:9525859
- ↑ Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003 May 1;423(6935):33-41. PMID:12721618 doi:http://dx.doi.org/10.1038/nature01580
- ↑ Waters MF, Minassian NA, Stevanin G, Figueroa KP, Bannister JP, Nolte D, Mock AF, Evidente VG, Fee DB, Muller U, Durr A, Brice A, Papazian DM, Pulst SM. Mutations in voltage-gated potassium channel KCNC3 cause degenerative and developmental central nervous system phenotypes. Nat Genet. 2006 Apr;38(4):447-51. Epub 2006 Feb 26. PMID:16501573 doi:ng1758
- ↑ 5.0 5.1 5.2 5.3 Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007 Nov 15;450(7168):376-82. PMID:18004376 doi:http://dx.doi.org/10.1038/nature06265
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Joel L. Sussman, Alexander Berchansky, Ann Taylor