Peroxisome Proliferator-Activated Receptors
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
===Co-Activator & Co-Repressor Binding=== | ===Co-Activator & Co-Repressor Binding=== | ||
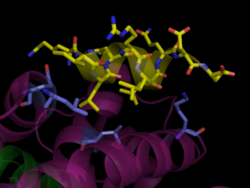
[[Image: SRC_binding.png|250px|left| Human PPARγ Co-Activator Binding Site. PPARγ bound to SRC-1, [[3dzy]]]] | [[Image: SRC_binding.png|250px|left| Human PPARγ Co-Activator Binding Site. PPARγ bound to SRC-1, [[3dzy]]]] | ||
| + | {{Clear}} | ||
The transcriptional activity of <scene name='Peroxisome_Proliferator-Activated_Receptors/Ppar_opening_2/2'>PPAR </scene>is regulated by its interaction with co-activators like SRC-1 or CBP and co-repressors like SMRT. <ref name="Zoete">PMID:17317294</ref>Co-activators like CBP contain a conserved LXXLL motif where X is any amino acid, and use this to bind a hydrophobic pocket on the receptor surface formed by the stabilized AF-2 helix H12.<ref name="Gampe">PMID:10882139</ref> In the case of the PPARγ/rosiglitazone/SRC-1 complex, the LXXLL motif helix of SRC-1 forms <scene name='Peroxisome_Proliferator-Activated_Receptors/Src_binding/1'>hydrophobic interactions with Leu468 and Leu318 of the LBD and hydrogen bonds between Glu471 and Lys301 and the co-activator backbone.</scene> These charged residues are conserved across PPAR isotypes and form the “charge clamp,” an essential component for co-activator stabilization in the PPAR LBD.<ref>PMID:11698662</ref> | The transcriptional activity of <scene name='Peroxisome_Proliferator-Activated_Receptors/Ppar_opening_2/2'>PPAR </scene>is regulated by its interaction with co-activators like SRC-1 or CBP and co-repressors like SMRT. <ref name="Zoete">PMID:17317294</ref>Co-activators like CBP contain a conserved LXXLL motif where X is any amino acid, and use this to bind a hydrophobic pocket on the receptor surface formed by the stabilized AF-2 helix H12.<ref name="Gampe">PMID:10882139</ref> In the case of the PPARγ/rosiglitazone/SRC-1 complex, the LXXLL motif helix of SRC-1 forms <scene name='Peroxisome_Proliferator-Activated_Receptors/Src_binding/1'>hydrophobic interactions with Leu468 and Leu318 of the LBD and hydrogen bonds between Glu471 and Lys301 and the co-activator backbone.</scene> These charged residues are conserved across PPAR isotypes and form the “charge clamp,” an essential component for co-activator stabilization in the PPAR LBD.<ref>PMID:11698662</ref> | ||
| Line 43: | Line 44: | ||
A number of synthetic agonists have been developed to bind to <scene name='Peroxisome_Proliferator-Activated_Receptors/Ppar_opening4/2'>PPAR</scene> to fight metabolic diseases like diabetes. These agonists include [http://en.wikipedia.org/wiki/troglitazone troglitazone] ([http://www.rezulin.com Rezulin]), pioglitazone ([[Actos]]), and rosiglitazone ([[Avandia]]). These agonists function in a similar fashion, by binding to the active site of PPARγ, activating the receptor. Rosiglitazone occupies roughly 40% of the LBD. It assumes a U-shaped conformation with the TZD head group forming a <scene name='Peroxisome_Proliferator-Activated_Receptors/Rosiglitazone_binding/3'>number of interactions that stabilize the agonist</scene>. Rosiglitazone forms hydrogen bond interactions with H323 and H449 and its TZD group, the sulfur atom of the TZD occupies a hydrophobic pocket formed by Phe363, Glu286, Phe282, Leu330, Ile326 and Leu469, and the central benzene ring occupies a pocket formed by Cys285 and Met364.<ref name="Nolte"/> | A number of synthetic agonists have been developed to bind to <scene name='Peroxisome_Proliferator-Activated_Receptors/Ppar_opening4/2'>PPAR</scene> to fight metabolic diseases like diabetes. These agonists include [http://en.wikipedia.org/wiki/troglitazone troglitazone] ([http://www.rezulin.com Rezulin]), pioglitazone ([[Actos]]), and rosiglitazone ([[Avandia]]). These agonists function in a similar fashion, by binding to the active site of PPARγ, activating the receptor. Rosiglitazone occupies roughly 40% of the LBD. It assumes a U-shaped conformation with the TZD head group forming a <scene name='Peroxisome_Proliferator-Activated_Receptors/Rosiglitazone_binding/3'>number of interactions that stabilize the agonist</scene>. Rosiglitazone forms hydrogen bond interactions with H323 and H449 and its TZD group, the sulfur atom of the TZD occupies a hydrophobic pocket formed by Phe363, Glu286, Phe282, Leu330, Ile326 and Leu469, and the central benzene ring occupies a pocket formed by Cys285 and Met364.<ref name="Nolte"/> | ||
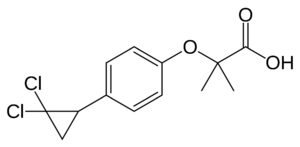
[[Image: Ciprofibrate.PNG|300px|left|thumb| Human PPARα agonist, Ciprofibrate (Modalim)]] | [[Image: Ciprofibrate.PNG|300px|left|thumb| Human PPARα agonist, Ciprofibrate (Modalim)]] | ||
| + | {{Clear}} | ||
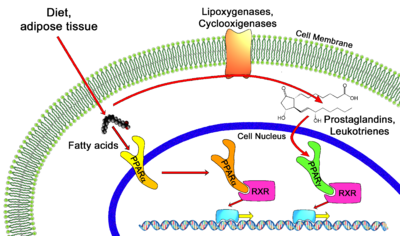
Despite their structural similarities, each member of the PPAR family is localized to certain parts of the body. Location of receptor partially determines their function in the body and also the different roles they can play in medicine as drug targets. PPARγ is responsible for lipid metabolism and cellular energy homeostasis. It binds genes that transcribe proteins which act as fatty acid transporters, are critical in insulin signaling and glucose transport, catalyze glycerol synthesis from triglycerides, and catabolize lipids. This makes PPARγ an ideal target to treat Diabetes.<ref name="Berger">PMID:11818483</ref> Also, recent research has indicated that some PPAR agonists like Rosiglitazone can induce apoptosis of macrophages and would thus serve as excellent anti-inflammatory targets.<ref name="Berger2">PMID:12079620</ref> PPARα has been shown to play a critical role in the regulation of uptake and oxidation of fatty acids. This makes PPARα an excellent target for Atherosclerosis drugs which aim to reduce LDL cholesterol and increase HDL cholesterol, the two most common traits of atherosclerosis. The fibrates are a class of amphipathic carboxylic acids that are PPARα agonists used to treat hypercholesterolemia and hyperlipidemia along with the [[HMGR]] inhibitor statins. Some fibrates are Bezafibrate (Marketed by Roche as [http://www.rxmed.com/b.main/b2.pharmaceutical/b2.1.monographs/CPS-%20Monographs/CPS-%20(General%20Monographs-%20B)/BEZALIP.html Bezalip]) and Ciprofibrate ([http://www.netdoctor.co.uk/medicines/100001714.html Modalim]).<ref name="Berger"/> PPARδ is broadly expressed across the human body and thus is suspected to play a role in a number of diseases. It has been implicated in disorders ranging from fertility problems to types of cancer. Perhaps the most important use of PPARδ agonists will be in treating central nervous system (CNS) diseases as PPARδ has been implicated in neuron myelinogenesis and neuronal signaling as well as lipid metabolism in the CNS.<ref name="Berger"/> | Despite their structural similarities, each member of the PPAR family is localized to certain parts of the body. Location of receptor partially determines their function in the body and also the different roles they can play in medicine as drug targets. PPARγ is responsible for lipid metabolism and cellular energy homeostasis. It binds genes that transcribe proteins which act as fatty acid transporters, are critical in insulin signaling and glucose transport, catalyze glycerol synthesis from triglycerides, and catabolize lipids. This makes PPARγ an ideal target to treat Diabetes.<ref name="Berger">PMID:11818483</ref> Also, recent research has indicated that some PPAR agonists like Rosiglitazone can induce apoptosis of macrophages and would thus serve as excellent anti-inflammatory targets.<ref name="Berger2">PMID:12079620</ref> PPARα has been shown to play a critical role in the regulation of uptake and oxidation of fatty acids. This makes PPARα an excellent target for Atherosclerosis drugs which aim to reduce LDL cholesterol and increase HDL cholesterol, the two most common traits of atherosclerosis. The fibrates are a class of amphipathic carboxylic acids that are PPARα agonists used to treat hypercholesterolemia and hyperlipidemia along with the [[HMGR]] inhibitor statins. Some fibrates are Bezafibrate (Marketed by Roche as [http://www.rxmed.com/b.main/b2.pharmaceutical/b2.1.monographs/CPS-%20Monographs/CPS-%20(General%20Monographs-%20B)/BEZALIP.html Bezalip]) and Ciprofibrate ([http://www.netdoctor.co.uk/medicines/100001714.html Modalim]).<ref name="Berger"/> PPARδ is broadly expressed across the human body and thus is suspected to play a role in a number of diseases. It has been implicated in disorders ranging from fertility problems to types of cancer. Perhaps the most important use of PPARδ agonists will be in treating central nervous system (CNS) diseases as PPARδ has been implicated in neuron myelinogenesis and neuronal signaling as well as lipid metabolism in the CNS.<ref name="Berger"/> | ||
Revision as of 12:33, 3 September 2015
| |||||||||||
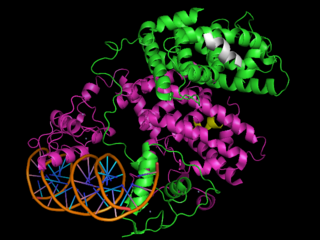
3D Structures of PPAR
Updated on 03-September-2015
Additional Resources
- See: Regulation of Gene Expression For Additional Mechanisms of Gene Regulation
- See: Pharmaceutical Drug Targets For Additional Information about Drug Targets for Related Diseases
- See: Diabetes & Hypoglycemia For Additional Information about Diabetes & Hypoglycemia Related Information
References
- ↑ 1.0 1.1 1.2 1.3 Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409-35. PMID:11818483 doi:10.1146/annurev.med.53.082901.104018
- ↑ Qi C, Zhu Y, Reddy JK. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem Biophys. 2000;32 Spring:187-204. PMID:11330046
- ↑ Guan HP, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 2005 Feb 15;19(4):453-61. Epub 2005 Jan 28. PMID:15681609 doi:10.1101/gad.1263305
- ↑ Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995 Jun;15(6):3012-22. PMID:7539101
- ↑ Lee SK, Jung SY, Kim YS, Na SY, Lee YC, Lee JW. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol Endocrinol. 2001 Feb;15(2):241-54. PMID:11158331
- ↑ Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999 Jun 25;284(5423):2174-7. PMID:10381882
- ↑ Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, Vidal-Puig A, Flier J, Briggs MR, Staels B, Vidal H, Auwerx J. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem. 1997 Jul 25;272(30):18779-89. PMID:9228052
- ↑ Girroir EE, Hollingshead HE, He P, Zhu B, Perdew GH, Peters JM. Quantitative expression patterns of peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) protein in mice. Biochem Biophys Res Commun. 2008 Jul 4;371(3):456-61. Epub 2008 Apr 28. PMID:18442472 doi:10.1016/j.bbrc.2008.04.086
- ↑ Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999 Jun 22;96(13):7473-8. PMID:10377439
- ↑ Gottlicher M, Widmark E, Li Q, Gustafsson JA. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4653-7. PMID:1316614
- ↑ Amri EZ, Bonino F, Ailhaud G, Abumrad NA, Grimaldi PA. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J Biol Chem. 1995 Feb 3;270(5):2367-71. PMID:7836471
- ↑ 12.0 12.1 12.2 Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998 Sep 10;395(6698):137-43. PMID:9744270 doi:10.1038/25931
- ↑ Fyffe SA, Alphey MS, Buetow L, Smith TK, Ferguson MA, Sorensen MD, Bjorkling F, Hunter WN. Recombinant human PPAR-beta/delta ligand-binding domain is locked in an activated conformation by endogenous fatty acids. J Mol Biol. 2006 Mar 3;356(4):1005-13. Epub 2006 Jan 4. PMID:16405912 doi:10.1016/j.jmb.2005.12.047
- ↑ Yang W, Rachez C, Freedman LP. Discrete roles for peroxisome proliferator-activated receptor gamma and retinoid X receptor in recruiting nuclear receptor coactivators. Mol Cell Biol. 2000 Nov;20(21):8008-17. PMID:11027271
- ↑ 15.0 15.1 15.2 15.3 15.4 Zoete V, Grosdidier A, Michielin O. Peroxisome proliferator-activated receptor structures: ligand specificity, molecular switch and interactions with regulators. Biochim Biophys Acta. 2007 Aug;1771(8):915-25. Epub 2007 Jan 18. PMID:17317294 doi:10.1016/j.bbalip.2007.01.007
- ↑ 16.0 16.1 16.2 Gampe RT Jr, Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu HE. Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000 Mar;5(3):545-55. PMID:10882139
- ↑ Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT Jr, McKee DD, Moore JT, Willson TM. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13919-24. Epub 2001 Nov 6. PMID:11698662 doi:10.1073/pnas.241410198
- ↑ Wahli W, Braissant O, Desvergne B. Peroxisome proliferator activated receptors: transcriptional regulators of adipogenesis, lipid metabolism and more.... Chem Biol. 1995 May;2(5):261-6. PMID:9383428
- ↑ Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008 Nov 20;456(7220):350-6. PMID:19043829 doi:10.1038/nature07413
- ↑ 20.0 20.1 Berger J, Wagner JA. Physiological and therapeutic roles of peroxisome proliferator-activated receptors. Diabetes Technol Ther. 2002;4(2):163-74. PMID:12079620
- ↑ http://uk.reuters.com/article/idUKT7482820080131
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Alexander Berchansky, Joel L. Sussman