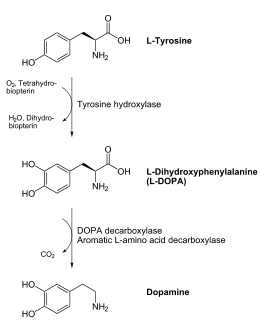
DOPA decarboxylase
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
{{Clear}} | {{Clear}} | ||
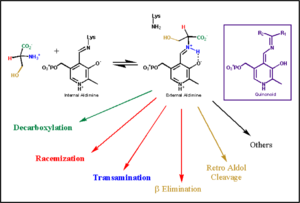
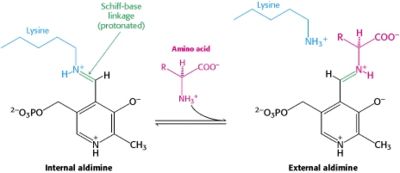
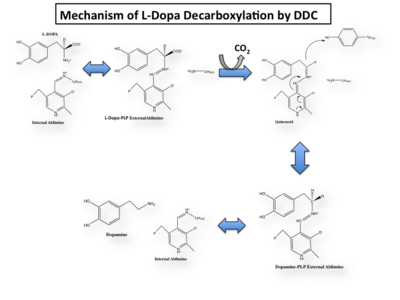
In the resting state, PLP forms a covalent bond with the amino group of the active site Lysine (internal aldimine). Upon introduction of the substrate into the active site, a new Schiff base is generated (external aldimine). This transimination step is common to '''all''' PLP-dependent enzymes. | In the resting state, PLP forms a covalent bond with the amino group of the active site Lysine (internal aldimine). Upon introduction of the substrate into the active site, a new Schiff base is generated (external aldimine). This transimination step is common to '''all''' PLP-dependent enzymes. | ||
| - | Although these enzymes have wide range of function, they can be classified into only five structural families: the '''aspartate amino transferase''', the tryptophan synthase β, the alanine racemase, the D-amino acid, and the glycogen phosphorylase. <ref name="percudani">PMID:12949584 </ref> <ref name="schneider">PMID:10673430 </ref> [[image:plp6.jpg|thumb|left|400px|'''The PLP is bound covalently to lysine residues in a Schiff base linkage (aldimine). In this form, it reacts with many free amino acids to replace the Schiff base to the lysine of the enzyme with a Schiff base to the amino acid substrate.'']] | + | Although these enzymes have wide range of function, they can be classified into only five structural families: the '''aspartate amino transferase''', the tryptophan synthase β, the alanine racemase, the D-amino acid, and the glycogen phosphorylase. <ref name="percudani">PMID:12949584 </ref> <ref name="schneider">PMID:10673430 </ref> [[image:plp6.jpg|thumb|left|400px|'''The PLP is bound covalently to lysine residues in a Schiff base linkage (aldimine). In this form, it reacts with many free amino acids to replace the Schiff base to the lysine of the enzyme with a Schiff base to the amino acid substrate.'']] |
| + | {{Clear}} | ||
---- | ---- | ||
===The Aspartate Aminotransferase Family=== | ===The Aspartate Aminotransferase Family=== | ||
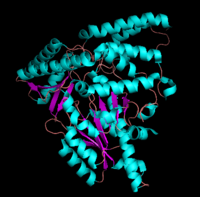
[[image:aspartateamino.png|thumb|left|200px|'''Aspartate Aminotransferase''']] | [[image:aspartateamino.png|thumb|left|200px|'''Aspartate Aminotransferase''']] | ||
| + | {{Clear}} | ||
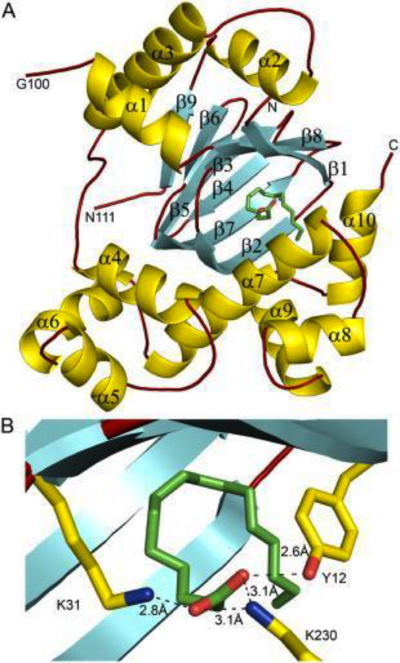
This family of PLP-dependent enzymes is also referred to as '''fold-type I''', with aspartate aminotransferase serving as the prototype. It is the most common structure of the five classes of PLP-dependent enzymes. This fold it is found in a variety of aminotransferases and decarboxylases, amongst them '''DOPA decarboxylase'''. PLP-dependent enzymes belonging to this family are catalytically active as homodimers and share a common, well-characterized structure, despite low-sequence identity. Each subunit has a large domain and a small domain. The central feature of the large domain is a seven-stranded β sheet. The small domain has either a three or four-stranded β sheet that is surrounded by α helices on one side. The cofactor PLP is covalently attached to a lysine residue in the large domain and is anchored in a way that allows the aromatic ring of PLP to pack against neighboring β strands. The active site is located in a cleft between the two domains at the interface between the two subunits. '''''Thus, enzymes of fold-type I have residues from both domains and both subunits involved in PLP-binding.''''' | This family of PLP-dependent enzymes is also referred to as '''fold-type I''', with aspartate aminotransferase serving as the prototype. It is the most common structure of the five classes of PLP-dependent enzymes. This fold it is found in a variety of aminotransferases and decarboxylases, amongst them '''DOPA decarboxylase'''. PLP-dependent enzymes belonging to this family are catalytically active as homodimers and share a common, well-characterized structure, despite low-sequence identity. Each subunit has a large domain and a small domain. The central feature of the large domain is a seven-stranded β sheet. The small domain has either a three or four-stranded β sheet that is surrounded by α helices on one side. The cofactor PLP is covalently attached to a lysine residue in the large domain and is anchored in a way that allows the aromatic ring of PLP to pack against neighboring β strands. The active site is located in a cleft between the two domains at the interface between the two subunits. '''''Thus, enzymes of fold-type I have residues from both domains and both subunits involved in PLP-binding.''''' | ||
---- | ---- | ||
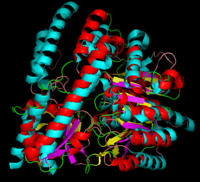
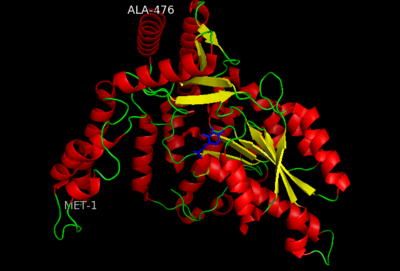
[[image:dopadecarb.png|thumb|200px|'''DOPA Decarboxylase''']][[image:super1.png|thumb|200px|'''DOPA decarboxylase superimposed on aspartate aminotransferase''']] | [[image:dopadecarb.png|thumb|200px|'''DOPA Decarboxylase''']][[image:super1.png|thumb|200px|'''DOPA decarboxylase superimposed on aspartate aminotransferase''']] | ||
| + | {{Clear}} | ||
---- | ---- | ||
===DOPA Decarboxylase=== | ===DOPA Decarboxylase=== | ||
| Line 48: | Line 51: | ||
---- | ---- | ||
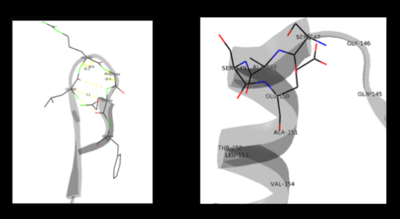
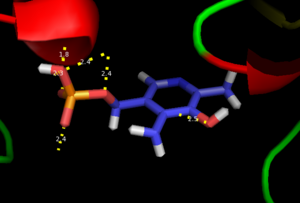
As well, a '''salt bridge''' exists between the carboxyl group of <scene name='DOPA_decarboxylase/Aspartic/1'>Asp271</scene> and the protonated pyridine nitrogen of PLP to further stabilize intermediate. Essentially, a salt bridge combines hydrogen bonding and electrostatic interactions (two common types non-covalent interactions). This interaction serves to provide an electron sink that can stabilize the carbanionic intermediates <ref name="jansonius">PMID:9914259 </ref> . PLP is further anchored to the protein by an extended '''hydrogen bond network''', as shown below. | As well, a '''salt bridge''' exists between the carboxyl group of <scene name='DOPA_decarboxylase/Aspartic/1'>Asp271</scene> and the protonated pyridine nitrogen of PLP to further stabilize intermediate. Essentially, a salt bridge combines hydrogen bonding and electrostatic interactions (two common types non-covalent interactions). This interaction serves to provide an electron sink that can stabilize the carbanionic intermediates <ref name="jansonius">PMID:9914259 </ref> . PLP is further anchored to the protein by an extended '''hydrogen bond network''', as shown below. | ||
| - | [[image:h-bonding.png|thumb|center|300px|'''H-bonding network of PLP in the active site''']] | + | [[image:h-bonding.png|thumb|center|300px|'''H-bonding network of PLP in the active site''']] |
| + | {{Clear}} | ||
---- | ---- | ||
The only two active site residues from the adjacent monomer, Ile-101 and Phe-103, are part of the substrate binding pocket. | The only two active site residues from the adjacent monomer, Ile-101 and Phe-103, are part of the substrate binding pocket. | ||
| Line 54: | Line 58: | ||
===Inhibitor Binding=== | ===Inhibitor Binding=== | ||
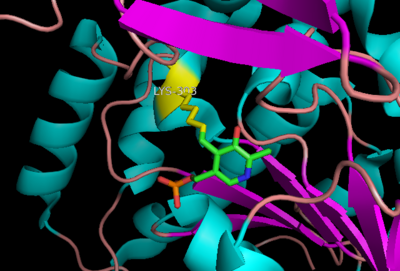
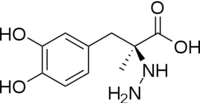
[[image:carbiDOPA.png|thumb|left|200px|'''carbiDOPA''']] | [[image:carbiDOPA.png|thumb|left|200px|'''carbiDOPA''']] | ||
| + | {{Clear}} | ||
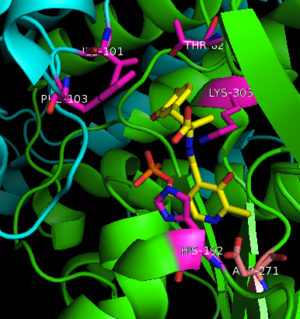
The inhibitor <scene name='DOPA_decarboxylase/Carbidopa/1'>carbiDOPA</scene> binds to the enzyme by forming a hydrazone linkage with PLP through its hydrazine moiety. The catechol ring of carbiDOPA is deeply buried in the active site cleft and is stabilized by <scene name='DOPA_decarboxylase/Vanderwaals/1'>van der waals contact</scene> with Ile-101 and Phe-103. The 4' hydroxyl group of the catechol ring participates in hydrogen bonding with <scene name='DOPA_decarboxylase/Thr-82/1'>Thr-82</scene>, further stabilizing the inhibitor in the active site cleft. PLP is further involved in substrate binding by forming a hydrogen bond to the 3' of the catechol ring. <scene name='DOPA_decarboxylase/His192/1'>His-192</scene>, a highly conserved residue of PLP-dependent decarboxylases <ref name="ishii">PMID:8889823 </ref> hydrogen bonds to the carboxylate group of carbiDOPA. | The inhibitor <scene name='DOPA_decarboxylase/Carbidopa/1'>carbiDOPA</scene> binds to the enzyme by forming a hydrazone linkage with PLP through its hydrazine moiety. The catechol ring of carbiDOPA is deeply buried in the active site cleft and is stabilized by <scene name='DOPA_decarboxylase/Vanderwaals/1'>van der waals contact</scene> with Ile-101 and Phe-103. The 4' hydroxyl group of the catechol ring participates in hydrogen bonding with <scene name='DOPA_decarboxylase/Thr-82/1'>Thr-82</scene>, further stabilizing the inhibitor in the active site cleft. PLP is further involved in substrate binding by forming a hydrogen bond to the 3' of the catechol ring. <scene name='DOPA_decarboxylase/His192/1'>His-192</scene>, a highly conserved residue of PLP-dependent decarboxylases <ref name="ishii">PMID:8889823 </ref> hydrogen bonds to the carboxylate group of carbiDOPA. | ||
[[image:actsite.png|thumb|center|300px|'''Key interactions between the active site residues, PLP, and carbiDOPA''']] | [[image:actsite.png|thumb|center|300px|'''Key interactions between the active site residues, PLP, and carbiDOPA''']] | ||
| - | + | {{Clear}} | |
'''Other inhibitors''' include [http://en.wikipedia.org/wiki/Benserazide Benserazide] (Serazide), α-Difluoromethyl-DOPA, and α-methyldopa. However, Benserazide is unapproved for use in the U.S. and is replaced by carbiDOPA instead. Both inhibitors act by preventing peripheral metabolism of L-DOPA, and cannot cross the blood-brain barrier. | '''Other inhibitors''' include [http://en.wikipedia.org/wiki/Benserazide Benserazide] (Serazide), α-Difluoromethyl-DOPA, and α-methyldopa. However, Benserazide is unapproved for use in the U.S. and is replaced by carbiDOPA instead. Both inhibitors act by preventing peripheral metabolism of L-DOPA, and cannot cross the blood-brain barrier. | ||
| Line 68: | Line 73: | ||
{{Clear}} | {{Clear}} | ||
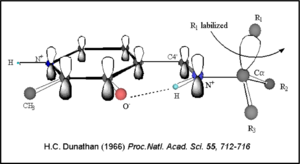
Once again, the transimination step (conversion of internal to external aldimine) is common to all PLP-dependent enzymes. The unique absorption wavelengths of the PLP intermediates has allowed for their straightforward detection (for example, an internal Schiff base in the enolimine form absorbs at 310-330 nm, whereas the quinonoid intermediate absorbs at ~500 nm). The orientation of the quinonoid intermediate allows for stereospecific decarboxylation of the substrate at the alpha carbon, as predicted by '''Dunathan's stereoelectronic hypothesis''' <ref name="dunathan">PMID:224217 </ref>, in which he proposed that the substrate binds PLP such that the external aldimine intermediate is oriented perpendicular to the coenzyme pi bonding system. In doing so, the sigma-pi orbital overlap in the transition state is maximized, thus maximizing the rate of the reaction. | Once again, the transimination step (conversion of internal to external aldimine) is common to all PLP-dependent enzymes. The unique absorption wavelengths of the PLP intermediates has allowed for their straightforward detection (for example, an internal Schiff base in the enolimine form absorbs at 310-330 nm, whereas the quinonoid intermediate absorbs at ~500 nm). The orientation of the quinonoid intermediate allows for stereospecific decarboxylation of the substrate at the alpha carbon, as predicted by '''Dunathan's stereoelectronic hypothesis''' <ref name="dunathan">PMID:224217 </ref>, in which he proposed that the substrate binds PLP such that the external aldimine intermediate is oriented perpendicular to the coenzyme pi bonding system. In doing so, the sigma-pi orbital overlap in the transition state is maximized, thus maximizing the rate of the reaction. | ||
| - | [[image:Dunathan.png|thumb|left|300px|'''Dunathan's Stereoeletronic Hypothesis, 1966''']] | + | [[image:Dunathan.png|thumb|left|300px|'''Dunathan's Stereoeletronic Hypothesis, 1966''']] |
| + | {{Clear}} | ||
This way, the developing p orbital is aligned for maximal overlap with the extended p system, lowering the energy of the transition state and increasing the rate of the reaction. As well, by controlling substrate orientation, the enzyme can distinguish between '''deprotonation''' and '''decarboxylation'''. | This way, the developing p orbital is aligned for maximal overlap with the extended p system, lowering the energy of the transition state and increasing the rate of the reaction. As well, by controlling substrate orientation, the enzyme can distinguish between '''deprotonation''' and '''decarboxylation'''. | ||
Revision as of 12:36, 3 September 2015
| |||||||||||
3D structures of DOPA decarboxylase
Updated on 03-September-2015
3k40 – DDC – Drosophila melanogaster
1js3 – pDDC + inhibitor – pig
1js6 - pDDC
3rbf, 3rbl – hDDC – human
3rch – hDDC + vitamin B6 phosphate + pyridoxal phosphate
References
- ↑ 1.0 1.1 Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000 Jan 15;8(1):R1-6. PMID:10673430
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003 Sep;4(9):850-4. PMID:12949584 doi:http://dx.doi.org/10.1038/sj.embor.embor914
- ↑ Maras B, Dominici P, Barra D, Bossa F, Voltattorni CB. Pig kidney 3,4-dihydroxyphenylalanine (dopa) decarboxylase. Primary structure and relationships to other amino acid decarboxylases. Eur J Biochem. 1991 Oct 15;201(2):385-91. PMID:1935935
- ↑ Aurora R, Rose GD. Helix capping. Protein Sci. 1998 Jan;7(1):21-38. PMID:9514257 doi:10.1002/pro.5560070103
- ↑ Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol. 1998 Dec;8(6):759-69. PMID:9914259
- ↑ 9.0 9.1 Ishii S, Mizuguchi H, Nishino J, Hayashi H, Kagamiyama H. Functionally important residues of aromatic L-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. J Biochem. 1996 Aug;120(2):369-76. PMID:8889823
- ↑ Hiscott JB, Defendi V. Simian virus 40 gene A regulation of cellular DNA synthesis. I. In permissive cells. J Virol. 1979 May;30(2):590-9. PMID:224217
- ↑ Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
Proteopedia Page Contributors and Editors (what is this?)
Brittany Todd, Michal Harel, David Canner, Alexander Berchansky, Brian Hernandez