Beta-secretase 1
From Proteopedia
(Difference between revisions)
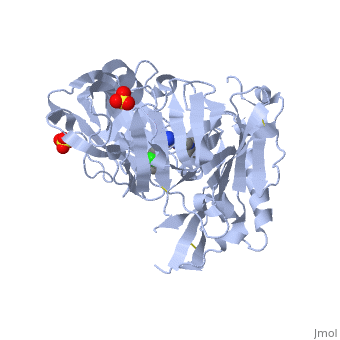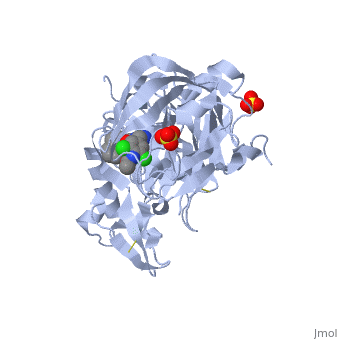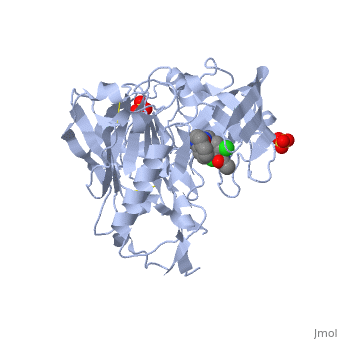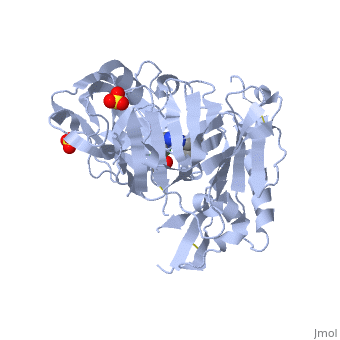
(New page: <StructureSection load='4ivt' size='340' side='right' caption='The best structure for Beta-secretase 1 shown: 4ivt' scene=''> Best example is PDB entry 4ivt and is shown in the viewer....) |
|||
| Line 1: | Line 1: | ||
| - | <StructureSection load='4ivt' size='340' side='right' caption='The best structure for Beta-secretase 1 shown: 4ivt' scene=''> | + | <StructureSection load='4ivt' size='340' side='right' caption='The best structure for Beta-secretase 1 shown: [[4ivt]]' scene=''> |
Best example is PDB entry [[4ivt]] and is shown in the viewer.<br> | Best example is PDB entry [[4ivt]] and is shown in the viewer.<br> | ||
Molecule Beta-secretase 1, also known as Beta-site amyloid precursor protein cleaving enzyme 1, Membrane-associated aspartic protease 2, Aspartyl protease 2, Asp 2, ASP2, Memapsin-2, Beta-secretase 1 and Beta-site APP cleaving enzyme 1. | Molecule Beta-secretase 1, also known as Beta-site amyloid precursor protein cleaving enzyme 1, Membrane-associated aspartic protease 2, Aspartyl protease 2, Asp 2, ASP2, Memapsin-2, Beta-secretase 1 and Beta-site APP cleaving enzyme 1. | ||
| Line 5: | Line 5: | ||
Responsible for the proteolytic processing of the amyloid precursor protein (APP). Cleaves at the N-terminus of the A-beta peptide sequence, between residues 671 and 672 of APP, leads to the generation and extracellular release of beta-cleaved soluble APP, and a corresponding cell-associated C-terminal fragment which is later released by gamma-secretase.Data source: Uniprot [http://www.uniprot.org/uniprot/P56817 P56817]<br> | Responsible for the proteolytic processing of the amyloid precursor protein (APP). Cleaves at the N-terminus of the A-beta peptide sequence, between residues 671 and 672 of APP, leads to the generation and extracellular release of beta-cleaved soluble APP, and a corresponding cell-associated C-terminal fragment which is later released by gamma-secretase.Data source: Uniprot [http://www.uniprot.org/uniprot/P56817 P56817]<br> | ||
==Catalytic Activity == | ==Catalytic Activity == | ||
| - | Broad endopeptidase specificity. Cleaves Glu-Val-Asn-Leu-|-Asp-Ala-Glu-Phe in the Swedish variant of Alzheimer amyloid precursor protein.Data source: Uniprot [http://www.uniprot.org/uniprot/P56817 P56817]<br> | + | Broad endopeptidase specificity. Cleaves Glu-Val-Asn-Leu-|-Asp-Ala-Glu-Phe in the Swedish variant of Alzheimer's amyloid precursor protein.Data source: Uniprot [http://www.uniprot.org/uniprot/P56817 P56817]<br> |
== Disease == | == Disease == | ||
| Line 16: | Line 16: | ||
proteolysis<br> | proteolysis<br> | ||
== In structures == | == In structures == | ||
| - | Beta-secretase 1 is found in | + | Beta-secretase 1 is found in 323 PDB entries<br> |
*<b>Eukaryota</b> (323 PDB entries): <br> | *<b>Eukaryota</b> (323 PDB entries): <br> | ||
| - | **<b>Homo sapiens</b> ( | + | **<b>Homo sapiens</b> (323 PDB entries): <br> |
***[[4ivt]] <div class="pdb-prints 4ivt"></div> | ***[[4ivt]] <div class="pdb-prints 4ivt"></div> | ||
***Title: Crystal structure of BACE1 with its inhibitor | ***Title: Crystal structure of BACE1 with its inhibitor | ||
Revision as of 20:20, 18 September 2015
| |||||||||||