Elizeu/sandbox/citocromo c
From Proteopedia
| Line 1: | Line 1: | ||
| - | == | + | ==Function== |
| - | + | [[Cas9]] is the RNA-guided DNA [[endonuclease]] used by the CRISPR (clustered regularly interspaced short palindromic repeats)-associated systems to generate double-strand DNA breaks in the invading DNA during an adaptive bacterial immune response. Three different types of CRISPR mechanisms have been discovered, however, only type II CRISPR systems are heavily researched. In vivo, Cas9 requires CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA)to guide the endonuclease toward invading DNA based on the complementary sequence recognition of these RNAs. Cas9 then targets and cleaves foreign DNA to interfere with viral replication. | |
| - | [[ | + | |
| + | The CRISPR-associated endonuclease Cas9 has been exploited for use in genome editing systems. In such systems, an engineered single-guide RNA (sgRNA) is used to perform the function of crRNA-tracRNA complex to target double-stranded breaks in genomic DNA. Depending on what repair pathway is triggered, often dictated by the inclusion of additional engineered components, the targeted site either is disrupted or incorporates additional genetic sequences. | ||
| - | + | [[Image:Layout for schematic and structure with structure.png|660px]] | |
| - | + | ==Interaction with DNA== | |
| + | Target DNA contains a protospacer adjacent motif on the non-complementary strand. This is a canonical sequence of 5’-NGG-3’ is recognized by Cas9 and is essential for genetic interference and editing, it will not cleave the target sequence if the PAM sequence is absent. The double Guanine of the non-complementary sequence strand interacts via hydrogen bonding from the major groove through two conserved arginine residues on the carboxy-terminus of Cas9(CITE #1). The minor groove of the PAM sequence interacts with a serine, through a hydrogen bridge to the last guanine, and lysine residue on the complementary target strand of the middle guanine. | ||
| - | + | The deoxyribose-phosphate backbone of the non-complementary strand is arranged in close proximity to various hydrogen bonding, some of which are accomplished through water molecules, and ionic interactions. (MMMOOORREE) | |
| - | + | ||
| - | + | ||
| - | + | Articles in Proteopedia concerning Cas9 include: | |
| + | * any? | ||
| - | ==References== | + | ==3D structures of Cas9== |
| + | |||
| + | ===''Streptococcus pyogenes'' Cas9=== | ||
| + | |||
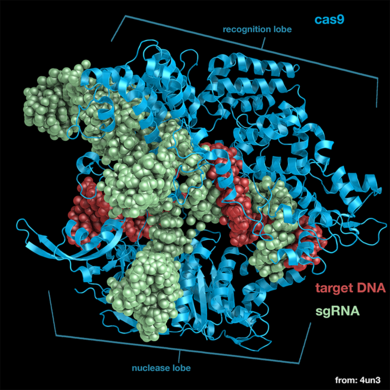
| + | * [[4un3]], [[4un4]], and [[4un5]] - ''S. pyogenes'' Cas9 bound to sgRNA and target DNA | ||
| + | [[Image:4un3 labeled.png|right|390px]] | ||
| + | * [[4oo8]] - ''S. pyogenes'' Cas9 bound to sgRNA and target DNA | ||
| + | * [[4cmp]] | ||
| + | * [[4cmq]] - Mn<sup>2+</sup>-bound ''S. pyogenes'' Cas9 | ||
| + | |||
| + | === ''Actinomyces naeslundii'' Cas9=== | ||
| + | *[[4oge]] | ||
| + | *[[4ogc]] - Mn<sup>2+</sup>-bound ''A.s naeslundii'' Cas9 | ||
| + | |||
| + | ==See Also== | ||
| + | [[HNH endonuclease]] <br/> | ||
| + | |||
| + | |||
| + | |||
| + | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | [[Category: Crispr]] | ||
| + | [[Category: Crispr-associated]] | ||
| + | [[Category: endonuclease]] | ||
Revision as of 20:03, 1 October 2015
Contents |
Function
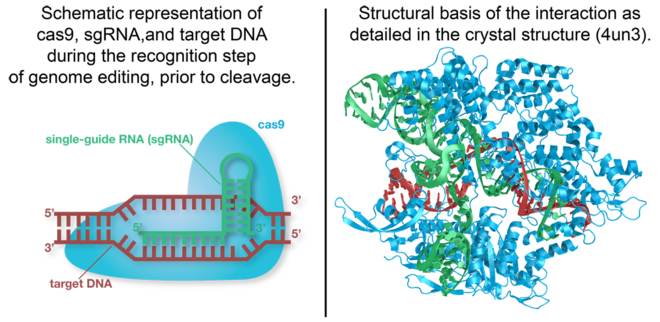
Cas9 is the RNA-guided DNA endonuclease used by the CRISPR (clustered regularly interspaced short palindromic repeats)-associated systems to generate double-strand DNA breaks in the invading DNA during an adaptive bacterial immune response. Three different types of CRISPR mechanisms have been discovered, however, only type II CRISPR systems are heavily researched. In vivo, Cas9 requires CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA)to guide the endonuclease toward invading DNA based on the complementary sequence recognition of these RNAs. Cas9 then targets and cleaves foreign DNA to interfere with viral replication.
The CRISPR-associated endonuclease Cas9 has been exploited for use in genome editing systems. In such systems, an engineered single-guide RNA (sgRNA) is used to perform the function of crRNA-tracRNA complex to target double-stranded breaks in genomic DNA. Depending on what repair pathway is triggered, often dictated by the inclusion of additional engineered components, the targeted site either is disrupted or incorporates additional genetic sequences.
Interaction with DNA
Target DNA contains a protospacer adjacent motif on the non-complementary strand. This is a canonical sequence of 5’-NGG-3’ is recognized by Cas9 and is essential for genetic interference and editing, it will not cleave the target sequence if the PAM sequence is absent. The double Guanine of the non-complementary sequence strand interacts via hydrogen bonding from the major groove through two conserved arginine residues on the carboxy-terminus of Cas9(CITE #1). The minor groove of the PAM sequence interacts with a serine, through a hydrogen bridge to the last guanine, and lysine residue on the complementary target strand of the middle guanine.
The deoxyribose-phosphate backbone of the non-complementary strand is arranged in close proximity to various hydrogen bonding, some of which are accomplished through water molecules, and ionic interactions. (MMMOOORREE)
Articles in Proteopedia concerning Cas9 include:
- any?
3D structures of Cas9
Streptococcus pyogenes Cas9
Actinomyces naeslundii Cas9
See Also
References
Proteopedia Page Contributors and Editors (what is this?)
Julie Langlois, Atena Farhangian, Rebecca Holstein, Elizabeth A. Dunlap, Katherine Reynolds, Elizeu Santos, Noam Gonen, Anna Lohning, Idan Ben-Nachum, Brian Ochoa, Shai Biran, Gauri Misra, Shira Weingarten-Gabbay, Keni Vidilaseris, Jamie Costa, Abhinav Mittal, Urs Leisinger, Madison Walberry, Edmond R Atalla, Brett M. Thumm, Brooke Fenn, Joel L. Sussman, Mati Cohen, Vesta Nwankwo, Dotan Shaniv, Gulalai Shah