Delta opioid receptor
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Background Information== | ==Background Information== | ||
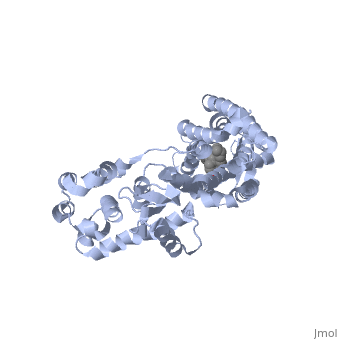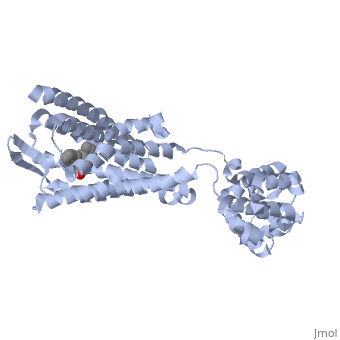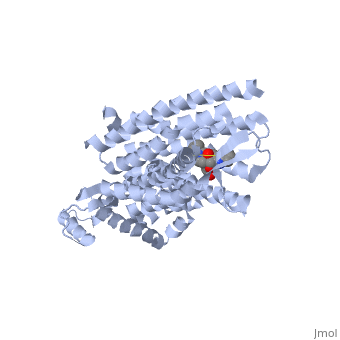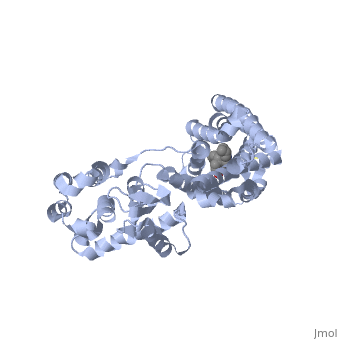
<StructureSection load='4ej4' size='340' side='right' caption='Cartoon representation of Delta opioid receptor bound to ligand in ball and stick representation.' scene=''> | <StructureSection load='4ej4' size='340' side='right' caption='Cartoon representation of Delta opioid receptor bound to ligand in ball and stick representation.' scene=''> | ||
| - | + | An opioid receptor is a membrane protein that acts as a receptor for endogenous neuropeptides called opioids. This receptor is an inhibitory G-protein coupled receptor that activates the parasympathetic nervous system in decreasing pain perception, slowing respiratory rate, improving mood, slowing digestion, and having an all-around analgesic effect on the body. Additionally, many agonists for this receptor are used as therapeutic drugs as powerful painkillers such as morphine, Percocet, and Vicodin. Some of these are naturally derived specifically from the poppy plant, and some are synthetically manufactured. There are three distinct opioid receptors, and the delta opioid receptor is one primarily responsible for the analgesic effects of opioids (Al-Hasani and Bruchas, 2011). | |
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | == | + | == Ligands == |
<scene name='71/715422/Sceneactivesite/1'>active site</scene> | <scene name='71/715422/Sceneactivesite/1'>active site</scene> | ||
The delta opioid receptor is shown with bound <scene name='71/715422/Sceneligand/1'>ligand</scene>. | The delta opioid receptor is shown with bound <scene name='71/715422/Sceneligand/1'>ligand</scene>. | ||
| - | == | + | == Mechanism of Action == |
Revision as of 22:34, 9 October 2015
Background Information
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644