Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 3'-sialyllactose
From Proteopedia
| Line 12: | Line 12: | ||
== Relevance == | == Relevance == | ||
| - | Influenza binds to sialic acid receptors present on the respiratory epithelium, eventually gaining access through endocytosis.<ref>Annu Rev Biochem</ref> By potentially concealing sialic acid receptors, it has been proposed that a sialic acid binding protein would aid in prevention of influenza infection. | + | Influenza binds to sialic acid receptors present on the respiratory epithelium, eventually gaining access through endocytosis.<ref>SkehelJJ, Wiley DC (2000) Receptor binding and Membrane fustion in virus entry: The influenza hemagglutinin. ''Annu Rev Biochem'' 69:531-569</ref> By potentially concealing sialic acid receptors, it has been proposed that a sialic acid binding protein would aid in prevention of influenza infection. <ref>Connaris H, Govorkova EA, Ligertwood Y, Dutia BM, Yang L, Tauber S, et al. Prevention of influenza by targeting host receptors using engineered proteins. Proc Natl Acad Sci U S A. 2014;111(17):6401–6406. '''[http://www.ncbi.nlm.nih.gov/pubmed/24733924?dopt=Abstract''' doi: 10.1073/pnas.1404205111''']'''</ref> |
== Structural highlights == | == Structural highlights == | ||
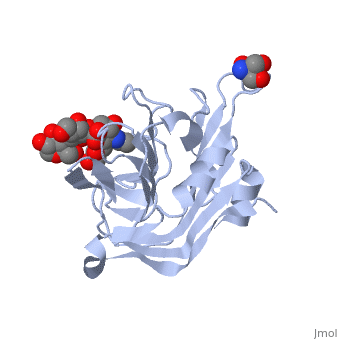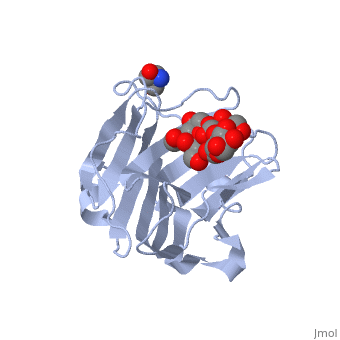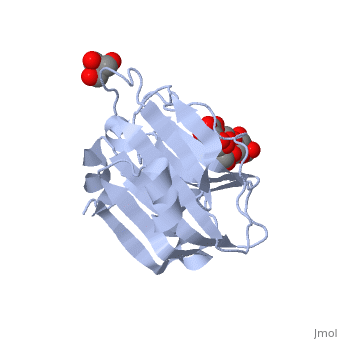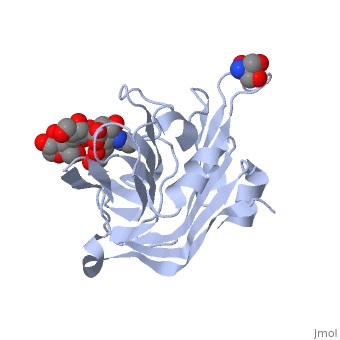
| - | <scene name='71/715437/Trs_site/1'>TRS site</scene> | + | The carbohydrate binding molecule (CBM) has a ligand chemical component identified with Tris(Hydroxymethyl)Aminomethane, an organic amine proton acceptor. It is used in the synthesis of surface-active agents and pharmaceuticals. This interaction can be see through the <scene name='71/715437/Trs_site/1'>TRS binding site</scene>. Lactose sialic acid is another identifier and ligand of the CBM <scene name='71/715437/Slt_binding/1'>SLT site</scene>. |
| - | <scene name='71/715437/Slt_binding/1'>SLT site</scene>. | + | Close interaction between lactose sialic acid and the carbohydrate binding molecule can be seen through <scene name='71/715437/Slt_binding_protein_space_fill/1'>SLT spacefill</scene> and <scene name='71/715437/Slt_binding_protein_spacefill/1'>SLT Spacefill Ligand</scene>. |
| - | <scene name='71/715437/Slt_binding_protein_space_fill/1'>SLT spacefill</scene> | + | |
| - | + | ||
| Line 26: | Line 24: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | <references/> | + | <references/> |
Revision as of 01:29, 10 October 2015
Contents |
Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 3'-sialyllactose
|
This is a default text for your page Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 3'-sialyllactose. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function
Designed to protect mice from H1N1 by masking the cell surface receptors recognized by influenza virion preventing viral binding and entry into the host cells.
Relevance
Influenza binds to sialic acid receptors present on the respiratory epithelium, eventually gaining access through endocytosis.[3] By potentially concealing sialic acid receptors, it has been proposed that a sialic acid binding protein would aid in prevention of influenza infection. [4]
Structural highlights
The carbohydrate binding molecule (CBM) has a ligand chemical component identified with Tris(Hydroxymethyl)Aminomethane, an organic amine proton acceptor. It is used in the synthesis of surface-active agents and pharmaceuticals. This interaction can be see through the . Lactose sialic acid is another identifier and ligand of the CBM . Close interaction between lactose sialic acid and the carbohydrate binding molecule can be seen through and .
</StructureSection>
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ SkehelJJ, Wiley DC (2000) Receptor binding and Membrane fustion in virus entry: The influenza hemagglutinin. Annu Rev Biochem 69:531-569
- ↑ Connaris H, Govorkova EA, Ligertwood Y, Dutia BM, Yang L, Tauber S, et al. Prevention of influenza by targeting host receptors using engineered proteins. Proc Natl Acad Sci U S A. 2014;111(17):6401–6406. doi: 10.1073/pnas.1404205111