Ivan Koutsopatriy estrogen receptor
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Structural highlights == | == Structural highlights == | ||
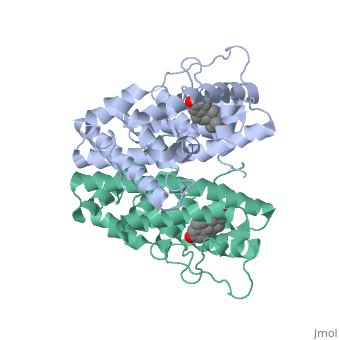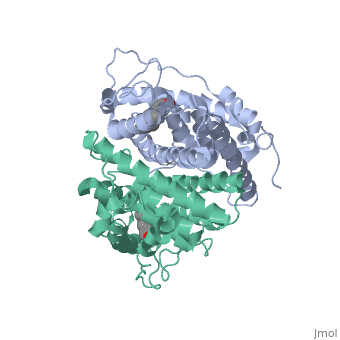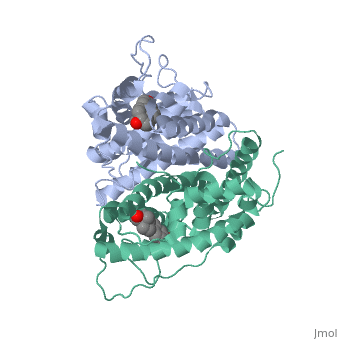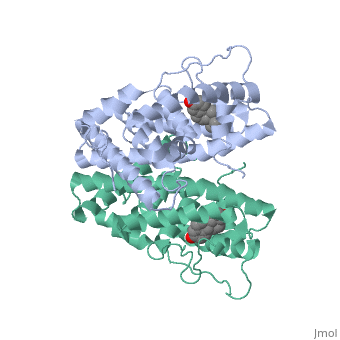
| - | ER is a modular protein composed of a ligand binding domain, a DNA binding domain and a transactivation domain. ER is a DNA-binding transcription factor. <scene name='71/714947/Er_bound_to_dna_dnadomain/1'> ER bound to DNA with one DNA binding helix and the transactivation domain highlighted yellow </scene> The DNA binding domain can be clearly observed in this scene; the highlighted yellow helix in close proximety to the DNA is part of the DNA binding domain. The blue beta sheet close to the yellow DNA binding alpha helix is also part of the DNA binding domain. The transactivation domain is attached at the end of the yellow DNA binding domain, also forming an alpha helix colored in yellow. The transactivation domain activates RNA polymerase when the receptor binds to DNA. The ligand binding domain may be observed here with the following scene. <scene name='71/714947/Agonist_ferutinine_bound_er/2'> Agonist_ferutinine_bound_er</scene> The ligand ferutinine is bound by the ligand binding domain, composed of | + | ER is a modular protein composed of a ligand binding domain, a DNA binding domain and a transactivation domain. ER is a DNA-binding transcription factor. <scene name='71/714947/Er_bound_to_dna_dnadomain/1'> ER bound to DNA with one DNA binding helix and the transactivation domain highlighted yellow </scene> The DNA binding domain can be clearly observed in this scene; the highlighted yellow helix in close proximety to the DNA is part of the DNA binding domain. The blue beta sheet close to the yellow DNA binding alpha helix is also part of the DNA binding domain. The transactivation domain is attached at the end of the yellow DNA binding domain, also forming an alpha helix colored in yellow. The transactivation domain activates RNA polymerase when the receptor binds to DNA. The ligand binding domain may be observed here with the following scene. <scene name='71/714947/Agonist_ferutinine_bound_er/2'> Agonist_ferutinine_bound_er</scene> The ligand ferutinine is bound by the ligand binding domain, composed of the surrounding blue colored alpha helices . Unbound ER normally exists loosly around the nucleus; this is subject to change depending on a multitude of factors including cell type, progress through cell cycle and reception of cellular signals. When estrogen enters the cell and binds ER, ER trans-locates and undergoes a conformation shift.<ref> Beato, M., Chavez, S., and Truss, M. (1996). Transcriptional regulation by steroid hormones. Steroids 61: 240–251. </ref> Ligand bound estrogen receptor associates more tightly with the nucleus. |
| Line 23: | Line 23: | ||
| - | ER is functional as a ligand-dependent transcription factor. <ref> Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG (25 May 2001)."Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity.". J Biol Chem. 276 (21): 18375–83. </ref> ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. Differences in the structure of the receptor are observed depending on what ligand ER has bound. Through comparisons of ER bound to agonist and antagonist ligands some structural components may be highlighted. <scene name='71/714947/Agonist_estradiol_bound_er/2'>Agonist_estradiol_bound_er</scene> The specific conformation of this tight loop creates part of the activation signal that will stimulate normal growth, as estradiol is a normal ligand for ER and allows for binding in the major groove of DNA. If the ligand is an antagonist the transcription factor function of estrogen receptor becomes hindered. <scene name='71/714947/Partial_agonist_genistein_er/3'>Partial_Agonist_genistein_bound_Er</scene> The conformation of ER bound to the partial agonist genistein has a loop which is not as tight as those found on ER with a complete agonist ligand. This is noticeable in the size difference of the pure agonist vs partial agonist scenes. | + | ER is functional as a ligand-dependent transcription factor. <ref> Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG (25 May 2001)."Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity.". J Biol Chem. 276 (21): 18375–83. </ref> ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. Differences in the structure of the receptor are observed depending on what ligand ER has bound. Through comparisons of ER bound to agonist and antagonist ligands some structural components may be highlighted. <scene name='71/714947/Agonist_estradiol_bound_er/2'>Agonist_estradiol_bound_er</scene> The specific conformation of this tight loop creates part of the activation signal that will stimulate normal growth, as estradiol is a normal ligand for ER and allows for binding in the major groove of DNA. If the ligand is an antagonist the transcription factor function of estrogen receptor becomes hindered. <scene name='71/714947/Partial_agonist_genistein_er/3'>Partial_Agonist_genistein_bound_Er</scene> The conformation of ER bound to the partial agonist genistein has a loop which is not as tight as those found on ER with a complete agonist ligand. This is noticeable in the size difference of the pure agonist vs partial agonist scenes. Special attention should be given to the bottom right alpha helices and beta sheets that are pushed out more in the antagonist compared to the agonist. |
Revision as of 01:35, 28 October 2015
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Beato, M., Chavez, S., and Truss, M. (1996). Transcriptional regulation by steroid hormones. Steroids 61: 240–251.
- ↑ Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG (25 May 2001)."Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity.". J Biol Chem. 276 (21): 18375–83.
- ↑ Htun H, Holth LT, Walker D, Davie JR, Hager GL (1 February 1999). "Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor". Mol Biol Cell 10 (2): 471–86.