Timothy Locksmith sandbox Heat Sock Factor
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Sturcture/DNA Interaction == | == Sturcture/DNA Interaction == | ||
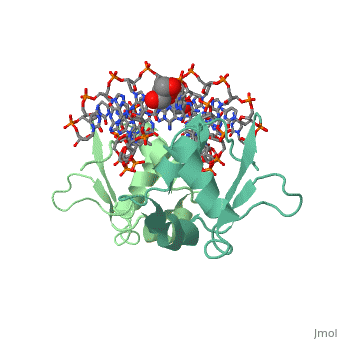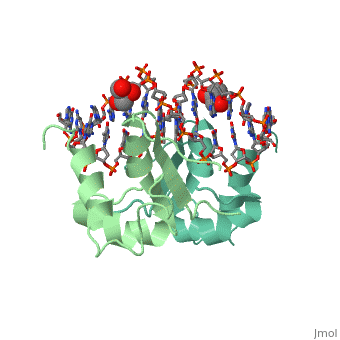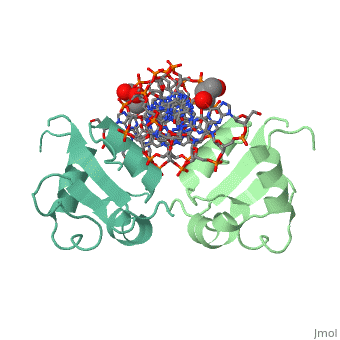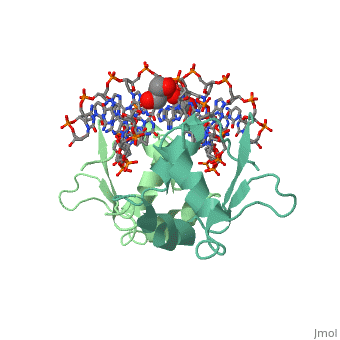
| - | HSF remains as a monomer when bound with Hsp under non-stressed conditions. Under stressed conditions (increased temperature) three individual monomers of HSF move into the cell’s nucleus, <scene name='71/714949/Dimer_hsf_with_dna_interaction/3'>trimerizes</scene> (could only isloate a dimer structure), and then bind to the large Groove of the DNA to Heat shock elements throughout the genome.<ref>DOI 10.1126/science.8421783</ref> Specifically an <scene name='71/714949/Arg_met_interact/1'>Arginine, and Methionine</scene> from each of the monomer bind to three oppositely oriented "<scene name='71/714949/Gaa_met_arg_interact/2'>nGAAn</scene>" sections of the genome. | + | HSF remains as a monomer when bound with Hsp under non-stressed conditions. Under stressed conditions (increased temperature) three individual monomers of HSF move into the cell’s nucleus, <scene name='71/714949/Dimer_hsf_with_dna_interaction/3'>trimerizes</scene> (could only isloate a dimer structure), and then bind to the large Groove of the DNA to Heat shock elements throughout the genome.<ref>DOI 10.1126/science.8421783</ref> Specifically an <scene name='71/714949/Arg_met_interact/1'>Arginine, and Methionine</scene> from each of the monomer bind to <scene name='71/714949/Alternating_gaa/1'>three oppositely oriented</scene> "<scene name='71/714949/Gaa_met_arg_interact/2'>nGAAn</scene>" sections of the genome. These residues interact with the nucleotide bases, while nearby residues interact with the back bone to stabilize the transcription factor in place. The structure is classified as a "winged" helix turn helix |
Revision as of 02:31, 28 October 2015
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Littlefield O, Nelson HC. A new use for the 'wing' of the 'winged' helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol. 1999 May;6(5):464-70. PMID:10331875 doi:10.1038/8269
- ↑ PMCID: PMC4011729
- ↑ doi: https://dx.doi.org/10.1126/science.8421783