Gunnar Reiske/Sandbox 102
From Proteopedia
m |
|||
| Line 32: | Line 32: | ||
== Treatments == | == Treatments == | ||
| - | + | Prolyl endopeptidases (PEPs) are a family of serine protease enzymes that help accelerate the breakdown of proline residues in peptides. Since gluten is a proline-rich complex, these enzymes can be used to help treat individuals with celiac disease. <ref> Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.</ref> | |
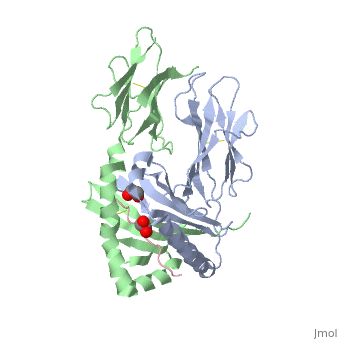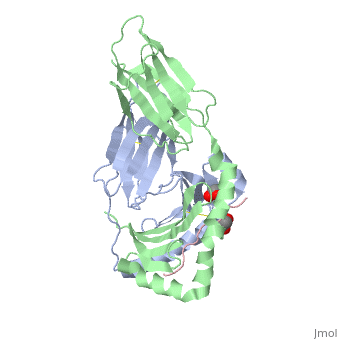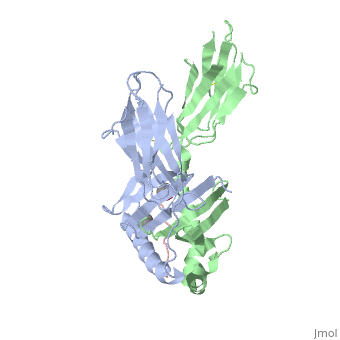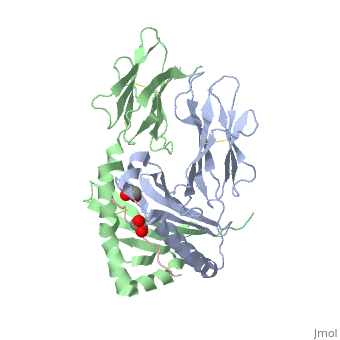
| - | + | A study was done to explore the structural features of two bacterial PEPs, one with a bound enzyme and one without (Figure 1). Both PEPs have two domains: one that is the catalytic binding site and one called the propeller domain. With further investigation of domain features of the PEP isolated from Myxococcus xanthus, interactions were observed that give the enzyme its proline-cleaving properties (Figure 2). | |
| + | |||
| + | Using the structural components of the open and bound forms of PEP enzymes, a mechanism was proposed in which the incoming proline-rich peptide causes a conformational change that opens the catalytic binding site. This conformation is stabilized by the prolines in the substrate interacting with the arginine and aspartate residues in the binding site. The propeller region does not interact with the bound substrate, but the aspartates and glutamate residues interact with arginine residues in the catalytic region to stabilize the unbound form of the enzyme. | ||
| + | |||
| + | Bacterial PEPs can detoxify immunotoxic proline-rich peptides in gut lumen of celiac patients by breaking down the gliadin before it reaches the small intestines. Given orally as a therapeutic treatment for those with celiac disease. Further research would need to be done on how we can make these enzymes more acid-stable to withstand the acidic environment of the human intestine. <ref>Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.</ref> | ||
| + | |||
| + | Studies have been done to determine the feasibility of the therapeutic implementation of bacterial PEP’s for detoxification of gliadin complexes in individuals with celiac disease. The study showed that a substantially high concentration of PEP’s as well as long exposure times (3 hours) were required for a complete detoxification of gliadin peptides and thus prevent intestinal transport of the peptides. <ref>Matysiak-Budnik, T., Candalh, C., Cellier, C., Dugave, C., Namane, A., Vidal-Martinez, T., . . . Heyman, M. (2005). Limited efficiency of prolyl-endopeptidase in the detoxification of gliadin peptides in celiac disease. Gastroenterology,129(3), 786-796. doi:10.1053/j.gastro.2005.06.016</ref> | ||
== Structural highlights == | == Structural highlights == | ||
Revision as of 00:28, 16 November 2015
How Gluten Protein Structure Stimulates an Immune Response
Introduction
The protein, gluten is found in wheat and grains such as rye and barley. Gluten is also involved with inducing an inflammatory response in individuals with celiac disease. Individuals who have the disease cannot digest gluten due to the protein’s structure, which will damage the small intestine. In detail, if an individual with celiac disease ingests foods containing gluten, the immune system responds by damaging the villi, which are fingerlike projections lining the small intestine. This type of immune response denies the body’s ability to absorb nutrients that pass through the small intestine and into the bloodstream. As a result of the damaged villi, people with celiac disease can become malnourished. Although celiac disease is genetic, the question of how the protein triggers an immune response in the gastrointestinal tract of affected individuals was further explored.
Gluten is a protein complex comprised of gliadin and glutenin. Gliadins, for those with celiac disease, are the principle toxic component of gluten and are composed of proline and glutamine peptide sequences. The peptides enter the circulatory system and come into contact with lymphocytes and T cells, resulting in the release of inflammatory chemicals. The inflammatory chemicals interact with the villi of the small intestine and damage them, disabling the body from nutrient absorption. The symptoms can include abdominal pain, weight loss, fatigue, and many other symptoms associated with malnutrition. As of now, the only treatment for celiac disease is the total exclusion of gluten from the person’s diet.[1]
| |||||||||||
References
- ↑ Celiac Disease: MedlinePlus. Retrieved October 27, 2015, from https://www.nlm.nih.gov/medlineplus/celiacdisease.html
- ↑ Luigi Maiuri, Carolina Ciacci, Ida Ricciardelli, Loredana Vacca, Valeria Raia, Salvatore Auricchio, Jean Picard, Mohamed Osman, Sonia Quaratino, Marco Londei, Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease, The Lancet, Volume 362, Issue 9377, 5 July 2003, Pages 30-37, ISSN 0140-6736, http://dx.doi.org/10.1016/S0140-6736(03)13803-2. (http://www.sciencedirect.com/science/article/pii/S0140673603138032)
- ↑ Luigi Maiuri, Carolina Ciacci, Ida Ricciardelli, Loredana Vacca, Valeria Raia, Salvatore Auricchio, Jean Picard, Mohamed Osman, Sonia Quaratino, Marco Londei, Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease, The Lancet, Volume 362, Issue 9377, 5 July 2003, Pages 30-37, ISSN 0140-6736, http://dx.doi.org/10.1016/S0140-6736(03)13803-2. (http://www.sciencedirect.com/science/article/pii/S0140673603138032)
- ↑ Kim, Quartsen, Bergsen, Khosla, & Sollid. (n.d.). Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Cross Mark, 101(12), 4175-4179. March 2004 http://www.pnas.org/content/101/12/4175.figures-only
- ↑ Kate N. Henderson, Jason A. Tye-Din, Hugh H. Reid, Zhenjun Chen, Natalie A. Borg, Tim Beissbarth, Arthur Tatham, Stuart I. Mannering, Anthony W. Purcell, Nadine L. Dudek, David A. van Heel, James McCluskey, Jamie Rossjohn, Robert P. Anderson, A Structural and Immunological Basis for the Role of Human Leukocyte Antigen DQ8 in Celiac Disease, Immunity, Volume 27, Issue 1, 27 July 2007, Pages 23-34, ISSN 1074-7613, http://dx.doi.org/10.1016/j.immuni.2007.05.015. (http://www.sciencedirect.com/science/article/pii/S1074761307003275)
- ↑ Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.
- ↑ Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.
- ↑ Matysiak-Budnik, T., Candalh, C., Cellier, C., Dugave, C., Namane, A., Vidal-Martinez, T., . . . Heyman, M. (2005). Limited efficiency of prolyl-endopeptidase in the detoxification of gliadin peptides in celiac disease. Gastroenterology,129(3), 786-796. doi:10.1053/j.gastro.2005.06.016
Proteopedia Page Contributors and Editors (what is this?)
Ben Horansky, Devin Joseph, Premal Patel, Gunnar Reiske, Katlin Cannon