Aminopeptidase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
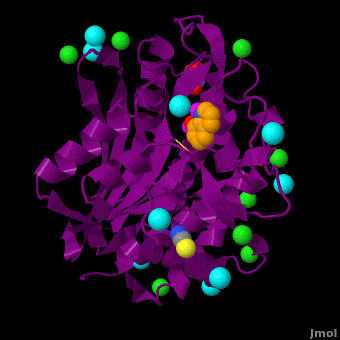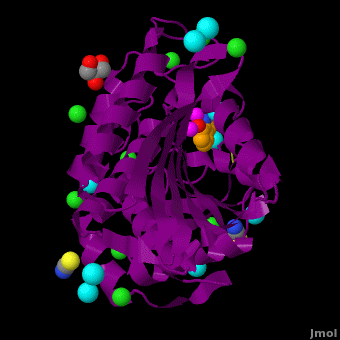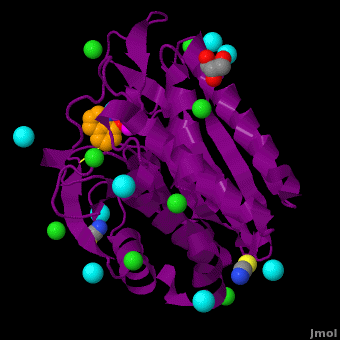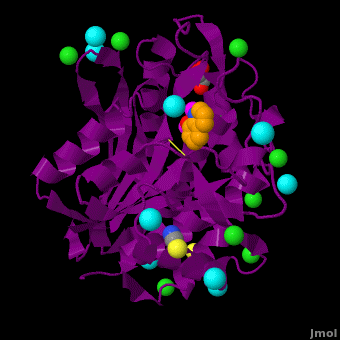
| - | <StructureSection load='VH9.pdb' size=' | + | <StructureSection load='VH9.pdb' size='350' side='right' scene='Journal:JBIC:15/Cv/1' caption='Bacterial leucine aminopeptidase complex with 8-hydroxyquinoline, glycerol, SCN, Zn+2 (magenta), Na+ (cyan) and Cl- (green) ions (PDB code [[3vh9]])'>[[Aminopeptidase|Aminopeptidases]] (AP) ([[EC]] 3) are metal - mostly Zn-dependent enzymes involved in the digestion of proteins. Cytosol AP (Cyt-AP), deblocking AP (DAP) and AP N (APN) remove N-terminal amino acids. The AP are classified by the amino acid which they hydrolyze. Other types of AP are:<br /> |
* '''Cold-activated AP''' (Col-AP)<br /> | * '''Cold-activated AP''' (Col-AP)<br /> | ||
* '''Heat stable AP''' from ''Thermus thermophilus'' (AmpT)<br /> | * '''Heat stable AP''' from ''Thermus thermophilus'' (AmpT)<br /> | ||
Revision as of 09:25, 2 December 2015
| |||||||||||
3D Structures of Aminopeptidase
Updated on 02-December-2015
4pv4]] – Pro-AP II + Mg – Yersinia pestis
Additional Resources
For additional information, see:
Amino Acid Synthesis & Metabolism
Streptomyces griseus Aminopeptidase (SGAP)
References
- ↑ Hanaya K, Suetsugu M, Saijo S, Yamato I, Aoki S. Potent inhibition of dinuclear zinc(II) peptidase, an aminopeptidase from Aeromonas proteolytica, by 8-quinolinol derivatives: inhibitor design based on Zn(2+) fluorophores, kinetic, and X-ray crystallographic study. J Biol Inorg Chem. 2012 Feb 5. PMID:22311113 doi:10.1007/s00775-012-0873-4
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, Eran Hodis