Sanbox glut3
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
==Structure== | ==Structure== | ||
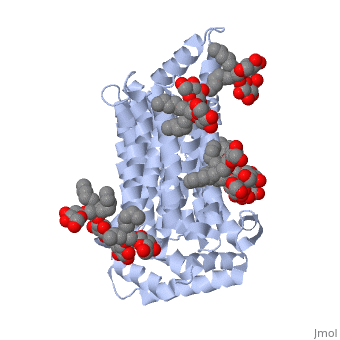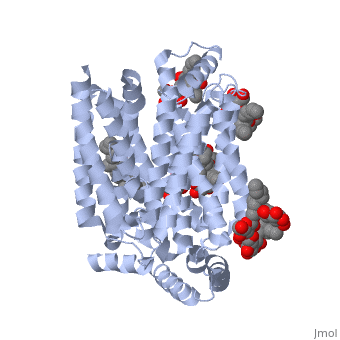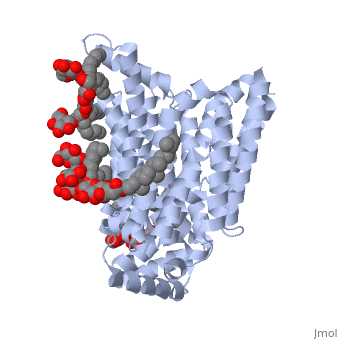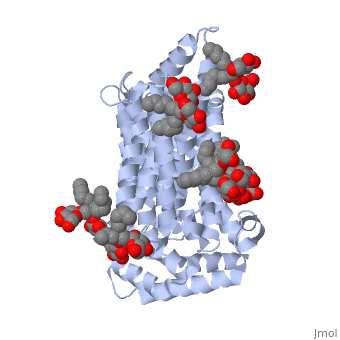
GLUT3(<scene name='71/716527/5c65/1'>5c65</scene>) is a transport protein consisting of 481 amino acids and weighing 52,520 Daltons in its asymmetrical unit<ref name="nineteen">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/</ref>. This protein is an alpha-helical protein consisting of two chains, two different ligands and water<ref name="nineteen"/>. The structure was determined by X-Ray diffraction and was measured at a resolution of 2.65 Angstroms<ref name="twentytwo">http://oca.weizmann.ac.il/oca-bin/ocaids?id=5c65</ref>. GLUT3 consists of 12 transmembrane segments (TMs) folded “into the N-terminal and C-terminal domains, each comprising ‘3+3’ inverted repeats”<ref name="nine"/> These TMs consist of four 3 repeated sections. [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F1.html Here] is a figure by Deng, D., et al. showing these repeated transmembrane segments<ref name="nine">Deng, D., Sun, P., Yan, C., Ke, M., Jiang, X., Xiong, L., . . . Yan, N. (2015). Molecular basis of ligand recognition and transport by glucose transporters. Nature, 526(7573), 391-396. doi:10.1038/nature14655</ref>. The protein consists of two different ligands, Y01 and 37X<ref name="eighteen">http://www.rcsb.org/pdb/explore.do?structureId=5C65</ref>. Octyl Glucose Neopentyl Glycol (<scene name='71/716527/37x/5'>37X</scene>) has a chemical formula of C<sub>27</sub>H<sub>52</sub>O<sub>12</sub> and a molecular weight of 569 Da. There are six 37X (501-506a) bound to chain A of 5c65. These ligands are kept in place by hydrogen bonds to arginine, proline, and serine and by van der Waals forces. Chain B has three 37X ligands attached to it (501-503b). These are attached through hydrogen bonds by arginine, proline, and serine as well as by van der Waals forces<ref name="twenty">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/37X</ref>. To view 37X in 3D use [http://www.rcsb.org/pdb/explore/jmol.do?structureId=5C65&residueNr=37X JSmol]. Cholesterol hemisuccinate (<scene name='71/716527/Y01/3'>Y01</scene>) has a chemical formula of C<sub>31</sub>H<sub>50</sub>O<sub>4</sub> and has a molecular weight of 487 Da. One Y01 is attached to chain a and another Y01 is attached to chain b<ref name="twentyone">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/Y01</ref>. To view Y01 in 3D use [http://www.rcsb.org/pdb/explore/jmol.do?structureId=5C65&residueNr=Y01 JSmol]. GLUT3 was also identified and analyzed in a complex with alpha & beta d-glucose. This model was reported with a resolution of 1.5 Å and was in an open-occluded state<ref name="nine"/>. The alpha and beta d glucose were coordinated in a <scene name='71/716527/Binding_pocket/1'>binding pocket</scene> by amino acids N315, E378, Q159, W368, Q280, Q281, N286. These are located on TM8 and TM10a and TM10b<ref name="nine"/>. A figure of this glucose coordination by Deng, D., et al. is available [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F2.html here]. GLUT3 structure was also determined when bound to maltose in an outward-open and an outward-occluded conformation. This was measure to a resolution of 2.6 Å and 2.4 Å respectively. A figure of this maltose coordination by Deng, D., et al. is available [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F2.html here]. To get a better view of the structure of the protein use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5C65 FirstGlance]. | GLUT3(<scene name='71/716527/5c65/1'>5c65</scene>) is a transport protein consisting of 481 amino acids and weighing 52,520 Daltons in its asymmetrical unit<ref name="nineteen">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/</ref>. This protein is an alpha-helical protein consisting of two chains, two different ligands and water<ref name="nineteen"/>. The structure was determined by X-Ray diffraction and was measured at a resolution of 2.65 Angstroms<ref name="twentytwo">http://oca.weizmann.ac.il/oca-bin/ocaids?id=5c65</ref>. GLUT3 consists of 12 transmembrane segments (TMs) folded “into the N-terminal and C-terminal domains, each comprising ‘3+3’ inverted repeats”<ref name="nine"/> These TMs consist of four 3 repeated sections. [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F1.html Here] is a figure by Deng, D., et al. showing these repeated transmembrane segments<ref name="nine">Deng, D., Sun, P., Yan, C., Ke, M., Jiang, X., Xiong, L., . . . Yan, N. (2015). Molecular basis of ligand recognition and transport by glucose transporters. Nature, 526(7573), 391-396. doi:10.1038/nature14655</ref>. The protein consists of two different ligands, Y01 and 37X<ref name="eighteen">http://www.rcsb.org/pdb/explore.do?structureId=5C65</ref>. Octyl Glucose Neopentyl Glycol (<scene name='71/716527/37x/5'>37X</scene>) has a chemical formula of C<sub>27</sub>H<sub>52</sub>O<sub>12</sub> and a molecular weight of 569 Da. There are six 37X (501-506a) bound to chain A of 5c65. These ligands are kept in place by hydrogen bonds to arginine, proline, and serine and by van der Waals forces. Chain B has three 37X ligands attached to it (501-503b). These are attached through hydrogen bonds by arginine, proline, and serine as well as by van der Waals forces<ref name="twenty">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/37X</ref>. To view 37X in 3D use [http://www.rcsb.org/pdb/explore/jmol.do?structureId=5C65&residueNr=37X JSmol]. Cholesterol hemisuccinate (<scene name='71/716527/Y01/3'>Y01</scene>) has a chemical formula of C<sub>31</sub>H<sub>50</sub>O<sub>4</sub> and has a molecular weight of 487 Da. One Y01 is attached to chain a and another Y01 is attached to chain b<ref name="twentyone">http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/Y01</ref>. To view Y01 in 3D use [http://www.rcsb.org/pdb/explore/jmol.do?structureId=5C65&residueNr=Y01 JSmol]. GLUT3 was also identified and analyzed in a complex with alpha & beta d-glucose. This model was reported with a resolution of 1.5 Å and was in an open-occluded state<ref name="nine"/>. The alpha and beta d glucose were coordinated in a <scene name='71/716527/Binding_pocket/1'>binding pocket</scene> by amino acids N315, E378, Q159, W368, Q280, Q281, N286. These are located on TM8 and TM10a and TM10b<ref name="nine"/>. A figure of this glucose coordination by Deng, D., et al. is available [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F2.html here]. GLUT3 structure was also determined when bound to maltose in an outward-open and an outward-occluded conformation. This was measure to a resolution of 2.6 Å and 2.4 Å respectively. A figure of this maltose coordination by Deng, D., et al. is available [http://www.nature.com/nature/journal/v526/n7573/fig_tab/nature14655_F2.html here]. To get a better view of the structure of the protein use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=5C65 FirstGlance]. | ||
| - | [[Image:http://www.nature.com/nature/journal/v526/n7573/images/nature14655-f2.jpg]] | ||
This is 5c65 shown with <scene name="/12/3456/Sample/1">colored groups</scene>. This is 5c65 shown as a <scene name="/12/3456/Sample/2">transparent representation</scene> of the protein. | This is 5c65 shown with <scene name="/12/3456/Sample/1">colored groups</scene>. This is 5c65 shown as a <scene name="/12/3456/Sample/2">transparent representation</scene> of the protein. | ||
Revision as of 03:59, 8 December 2015
Facilitated Glucose Transporter 3, Solute Carrier Family 2 (GLUT3/ SLC2A3) in Homo Sapiens
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Long, W., & Cheeseman, C. I. (2015). Structure of, and functional insight into the GLUT family of membrane transporters. Cell Health and Cytoskeleton, 7, 167-183. doi:10.2147/CHC.S60484
- ↑ 2.0 2.1 Kipmen-Korgun, D., Bilmen-Sarikcioglu, S., Altunbas, H., Demir, R., & Korgun, E. T. (2009). Type-2 diabetes down-regulates glucose transporter proteins and genes of the human blood leukocytes.Scandinavian Journal of Clinical and Laboratory Investigation, 69(3), 350-358. doi:10.1080/00365510802632163
- ↑ 3.0 3.1 Simpson,I. A., Dwyer, D., Malide, D., Moley, K. H., Travis, A., & Vannucci, S. J. (2008). The facilitative glucose transporter GLUT3: 20 years of distinction. American Journal of Physiology - Endocrinology and Metabolism, 295(2), E242-E253. doi:10.1152/ajpendo.90388.2008
- ↑ Maher, F., Vannucci, S. J., & Simpson, I. A. (1994). Glucose transporter proteins in brain. FASEB Journal, 8(13), 1003-1011.
- ↑ Xu, J., Lu, C., Wang, J., Zhang, R., Qian, X., & Zhu, H. (2015). Regulation of human trophoblast GLUT3 glucose transporter by mammalian target of rapamycin signaling. International Journal of Molecular Sciences, 16(6), 13815-13828. doi:10.3390/ijms160613815
- ↑ 6.0 6.1 Liu, Y., Liu, F., Iqbal, K., Grundke-Iqbal, I., & Gong, C. -. (2008). Decreased glucose transporters correlate to abnormal hyperphosphorylation of tau in alzheimer disease. FEBS Letters, 582(2), 359-364. doi:10.1016/j.febslet.2007.12.035
- ↑ 7.0 7.1 http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/
- ↑ http://oca.weizmann.ac.il/oca-bin/ocaids?id=5c65
- ↑ 9.0 9.1 9.2 9.3 Deng, D., Sun, P., Yan, C., Ke, M., Jiang, X., Xiong, L., . . . Yan, N. (2015). Molecular basis of ligand recognition and transport by glucose transporters. Nature, 526(7573), 391-396. doi:10.1038/nature14655
- ↑ http://www.rcsb.org/pdb/explore.do?structureId=5C65
- ↑ http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/37X
- ↑ http://www.ebi.ac.uk/pdbe/entry/pdb/5c65/bound/Y01
- ↑ Vittori, A., Breda, C., Repici, M., Orth, M., Roos, R. A. C., Outeiro, T. F., . . . the REGISTRY investigators of the European Huntington's Disease Network. (2014). Copy-number variation of the neuronal glucose transporter gene SLC2A3 and age of onset in huntington's disease. Human Molecular Genetics, 23(12), 3129-3137. doi:10.1093/hmg/ddu022
- ↑ McClory, H., Williams, D., & Sapp, E. (2014). Glucose transporter 3 is a rab11-dependent trafficking cargo and its transport to the cell surface is reduced in neurons of CAG140 Huntington’s disease mice. Acta Neuropathol Commun, 2, 1-9.
- ↑ 15.0 15.1 Naftalin RJ, Holman GD. Transport of sugars in human red cells. In: Ellory JC, Lew V, editors. \ Membrane Transport in Red Cells. New York, NY, USA: Academic Press; 1977.
- ↑ 16.0 16.1 Carruthers, A., DeZutter, J., Ganguly, A., & Devaskar, S. U. (2009). Will the original glucose transporter isoform please stand up! American Journal of Physiology - Endocrinology and Metabolism, 297(4), E836-E848. doi:10.1152/ajpendo.00496.2009
- ↑ Jardetzky, O. (1966). Simple allosteric model for membrane pumps [27]. Nature, 211(5052), 969-970. doi:10.1038/211969a0
- ↑ Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615.
- ↑ Caulfield MJ, Munroe PB, O’Neill D, et al. SLC2A9 is a high-capacity urate transporter in humans. PLoS Med. 2008;5:1509–1523.
- ↑ Vollers, S. S., & Carruthers, A. (2012). Sequence determinants of GLUT1-mediated accelerated-exchange transport: Analysis by homology-scanning mutagenesis. Journal of Biological Chemistry, 287(51), 42533-42544.doi:10.1074/jbc.M112.369587