Sandbox sortases
From Proteopedia
| Line 3: | Line 3: | ||
==Sortase System== | ==Sortase System== | ||
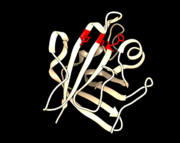
<StructureSection load='1T2P_A_sortaseA.pdb' size='340' side='right' caption='Class A sortase' scene=''> | <StructureSection load='1T2P_A_sortaseA.pdb' size='340' side='right' caption='Class A sortase' scene=''> | ||
| - | Sortase enzymes are trans-peptidases found in Gram-positive bacterial species. Their purpose is to covalently link proteins to the cell wall. By recognizing a specific sequence on target proteins, they “sort” which proteins to attach. Different sortases are separated into different classes based on their recognition sequence and specific function. Class A sortases (SrtA) found in ''Staphylococcus aureus'' were the first sortase enzyme to be isolated in the lab in 1999 and have become the prototypical sortase | + | Sortase enzymes are trans-peptidases found in Gram-positive bacterial species. Their purpose is to covalently link proteins to the cell wall. By recognizing a specific sequence on target proteins, they “sort” which proteins to attach. Different sortases are separated into different classes based on their recognition sequence and specific function. Class A sortases (SrtA) found in ''Staphylococcus aureus'' were the first sortase enzyme to be isolated in the lab in 1999 and have become the prototypical sortase <ref name= "Handbook of Proteolytic Enzymes">McCafferty, Dewey G., and Jeffrey A. Melvin. ‘Sortases’. Handbook of Proteolytic Enzymes. N.p.: Elsevier BV, 2013. 2459–2465. PDF.</ref>. Because surface proteins play such a big role in a pathogen’s virulence, sortases have become an important topic for study [2]. |
== Structure == | == Structure == | ||
| - | Sortases usually consist of 200-300 amino acids, with a typical molecular weight between 20-30 kDa. The enzyme possesses a hydrophobic transmembrane α-helix at the N-terminus, with the majority of the protein on the outside of the membrane. They have a typical “sortase-fold” that consists of an uneven β-barrel made up of eight β-strands. The connecting loops form the walls of a groove where the active site rests. It is these loops that are thought to give these enzymes their specificity | + | Sortases usually consist of 200-300 amino acids, with a typical molecular weight between 20-30 kDa. The enzyme possesses a hydrophobic transmembrane α-helix at the N-terminus, with the majority of the protein on the outside of the membrane. They have a typical “sortase-fold” that consists of an uneven β-barrel made up of eight β-strands. The connecting loops form the walls of a groove where the active site rests. It is these loops that are thought to give these enzymes their specificity <ref name= "Handbook of Proteolytic Enzymes" />. |
[[Image:1T2Pactivesite.png | thumb | PDB 1T2W Active Site Reisdues Arg 197, Cys 184, and His 120 highlighted]] | [[Image:1T2Pactivesite.png | thumb | PDB 1T2W Active Site Reisdues Arg 197, Cys 184, and His 120 highlighted]] | ||
== ''S. aureus'' SrtA mechanism == | == ''S. aureus'' SrtA mechanism == | ||
| - | In Gram-positive bacteria, SrtA enzymes are almost always present. Their prevalence, combined with them being discovered first, has led to them having the most extensive mechanistic studies of all the sortases. The active site is composed of an Arginine residue (Arg 197), a Cystine residue (Cys 184), and a Histidine residue (His 120). SrtA has a LPXTG recognition sequence that is highly conserved. Mobile loops peripheral to the active site bind to this sequence near the C-terminal. In ''S. aureus'' a calcium ion binds behind the substrate binding groove and locks the loops into a binding position. The Arg197 residue hydrogen bonds to the carbonyl on the peptide backbone of the substrate, meanwhile the Cys184 deprotonates into a thiolate ion to perform a nucleophilic attack on the bond between T and G on the substrate. The His120 residue donates a proton to the amine leaving group of the substrate’s C-terminal domain. Then an amine on the pentaglycine crossbridge protein of a peptidoglycan precursor enters the active site and has a proton abstracted by the histidine. This increases the amine’s nucleophilicity enabling a nucleophilic attack on substrate. This dislocates the substrate from the enzyme and restores the active site. This results in the two peptides bonded together. This process is entirely driven without spending ATP | + | In Gram-positive bacteria, SrtA enzymes are almost always present. Their prevalence, combined with them being discovered first, has led to them having the most extensive mechanistic studies of all the sortases. The active site is composed of an Arginine residue (Arg 197), a Cystine residue (Cys 184), and a Histidine residue (His 120). SrtA has a LPXTG recognition sequence that is highly conserved. Mobile loops peripheral to the active site bind to this sequence near the C-terminal. In ''S. aureus'' a calcium ion binds behind the substrate binding groove and locks the loops into a binding position. The Arg197 residue hydrogen bonds to the carbonyl on the peptide backbone of the substrate, meanwhile the Cys184 deprotonates into a thiolate ion to perform a nucleophilic attack on the bond between T and G on the substrate. The His120 residue donates a proton to the amine leaving group of the substrate’s C-terminal domain. Then an amine on the pentaglycine crossbridge protein of a peptidoglycan precursor enters the active site and has a proton abstracted by the histidine. This increases the amine’s nucleophilicity enabling a nucleophilic attack on substrate. This dislocates the substrate from the enzyme and restores the active site. This results in the two peptides bonded together. This process is entirely driven without spending ATP <ref name= "Handbook of Proteolytic Enzymes" />. |
== Sortase Classes == | == Sortase Classes == | ||
'''Class A''' | '''Class A''' | ||
| - | Sortase A enzymes, like SrtA, are known as “housekeeping” sortases. They recognize the sequence LPXTG and are nearly universally found in all Gram-positive bacteria. They sort and attach a wide variety of cell surface proteins to the cell wall | + | Sortase A enzymes, like SrtA, are known as “housekeeping” sortases. They recognize the sequence LPXTG and are nearly universally found in all Gram-positive bacteria. They sort and attach a wide variety of cell surface proteins to the cell wall <ref name= "Handbook of Proteolytic Enzymes" />. |
'''Class B''' | '''Class B''' | ||
| Line 22: | Line 22: | ||
'''Class C''' | '''Class C''' | ||
| - | Pili may extend 0.2-3.0 μm from the cell surface and promote cell adhesion and the formation of biofilms. In Gram-positive bacteria class C sortases are used to construct pili, linking together the pilin subunits via isopeptide bonds and only sometimes connects the pilus to the cell wall itself. The general process is conserved, but there is a greater variety in the structure or number of sortases and accessory factors needed [2]. The recognition sequence for class C sortases is QVPTG | + | Pili may extend 0.2-3.0 μm from the cell surface and promote cell adhesion and the formation of biofilms. In Gram-positive bacteria class C sortases are used to construct pili, linking together the pilin subunits via isopeptide bonds and only sometimes connects the pilus to the cell wall itself. The general process is conserved, but there is a greater variety in the structure or number of sortases and accessory factors needed [2]. The recognition sequence for class C sortases is QVPTG <ref name= "Handbook of Proteolytic Enzymes" />. |
'''Class D''' | '''Class D''' | ||
| - | Class D sortases have so far only been studied in ''B. anthracis'', where it was found to attach BasH and BasI to the cell wall. The class D sortase manages to attach each protein to different structures in the sporulating cell. Deleting the class D sortase reduced the efficiency of sporulation [2]. The recognition sequence for class D sortases is LPNTA | + | Class D sortases have so far only been studied in ''B. anthracis'', where it was found to attach BasH and BasI to the cell wall. The class D sortase manages to attach each protein to different structures in the sporulating cell. Deleting the class D sortase reduced the efficiency of sporulation [2]. The recognition sequence for class D sortases is LPNTA <ref name= "Handbook of Proteolytic Enzymes" />. |
'''Class E and F''' | '''Class E and F''' | ||
Revision as of 18:38, 11 December 2015
This page is setup for Brandon to build his senior project for OU CHEM 4923
Sortase System
| |||||||||||
References
1. McCafferty, Dewey G., and Jeffrey A. Melvin. ‘Sortases’. Handbook of Proteolytic Enzymes. N.p.: Elsevier BV, 2013. 2459–2465. PDF.
2. Spirig, T, EM Weiner, and RT Clubb. ‘Sortase Enzymes in Gram-Positive Bacteria’. Molecular microbiology. 5.82 (27 Oct. 2011): n.pag. 4 Nov. 2015.
3. Maresso, Anthony W., Travis J. Chapa, and Olaf Schneewind. ‘Surface Protein IsdC and Sortase B Are Required for Heme-Iron Scavenging of Bacillus Anthracis▿’. 188.23 (29 Sep. 2006): n.pag. 4 Nov. 2015.
4. Theile, Christopher S, et al. ‘Site-Specific N-Terminal Labeling of Proteins Using Sortase-Mediated Reactions’. Nature Protocols 8.9 (29 Aug. 2013): 1800–1807.
5. Mao, H, et al. ‘Sortase-Mediated Protein Ligation: A New Method for Protein Engineering’. Journal of the American Chemical Society. 9.126 (5 Mar. 2004): n.pag. 4 Nov. 2015.