Sandbox sortases
From Proteopedia
| Line 3: | Line 3: | ||
==Sortase System== | ==Sortase System== | ||
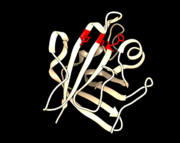
<StructureSection load='1T2P_A_sortaseA.pdb' size='340' side='right' caption='Class A sortase' scene=''> | <StructureSection load='1T2P_A_sortaseA.pdb' size='340' side='right' caption='Class A sortase' scene=''> | ||
| - | Sortase enzymes are trans-peptidases found in Gram-positive bacterial species. Their purpose is to covalently link proteins to the cell wall. By recognizing a specific sequence on target proteins, they “sort” which proteins to attach. Different sortases are separated into different classes based on their recognition sequence and specific function. Class A sortases (SrtA) found in ''Staphylococcus aureus'' were the first sortase enzyme to be isolated in the lab in 1999 and have become the prototypical sortase <ref name= "Handbook of Proteolytic Enzymes">McCafferty, Dewey G., and Jeffrey A. Melvin. ‘Sortases’. Handbook of Proteolytic Enzymes. N.p.: Elsevier BV, 2013. 2459–2465. PDF.</ref>. Because surface proteins play such a big role in a pathogen’s virulence, sortases have become an important topic for study | + | Sortase enzymes are trans-peptidases found in Gram-positive bacterial species. Their purpose is to covalently link proteins to the cell wall. By recognizing a specific sequence on target proteins, they “sort” which proteins to attach. Different sortases are separated into different classes based on their recognition sequence and specific function. Class A sortases (SrtA) found in ''Staphylococcus aureus'' were the first sortase enzyme to be isolated in the lab in 1999 and have become the prototypical sortase <ref name= "Handbook of Proteolytic Enzymes">McCafferty, Dewey G., and Jeffrey A. Melvin. ‘Sortases’. Handbook of Proteolytic Enzymes. N.p.: Elsevier BV, 2013. 2459–2465. PDF.</ref>. Because surface proteins play such a big role in a pathogen’s virulence, sortases have become an important topic for study <ref name= "spirig">Spirig, T, EM Weiner, and RT Clubb. ‘Sortase Enzymes in Gram-Positive Bacteria’. Molecular microbiology. 5.82 (27 Oct. 2011): n.pag. 4 Nov. 2015.</ref>. |
== Structure == | == Structure == | ||
| Line 18: | Line 18: | ||
'''Class B''' | '''Class B''' | ||
| - | The acquisition of iron plays an important role in the creation of bacterial infections and Class B sortases bind proteins that help sequester iron from the environment. SrtB is a class B sortase found in both ''S. aureus'' and ''Bacillus anthracis''. In both organsims they are associated with binding the protein IsdC to the cell wall which binds to heme and utilize it as a source of iron [3]. The sequence recognized by class B sortases is NP(Q/K)(T/S)(N/G/S)(D/A) displaying a wider variety of signals. Class B sortases noticeably attaches proteins to different sites in the cell wall than class A sortases. Another difference between SrtA and SrtB is that SrtB and the iron acquiring IsdC are only expressed under iron deficient conditions | + | The acquisition of iron plays an important role in the creation of bacterial infections and Class B sortases bind proteins that help sequester iron from the environment. SrtB is a class B sortase found in both ''S. aureus'' and ''Bacillus anthracis''. In both organsims they are associated with binding the protein IsdC to the cell wall which binds to heme and utilize it as a source of iron [3]. The sequence recognized by class B sortases is NP(Q/K)(T/S)(N/G/S)(D/A) displaying a wider variety of signals. Class B sortases noticeably attaches proteins to different sites in the cell wall than class A sortases. Another difference between SrtA and SrtB is that SrtB and the iron acquiring IsdC are only expressed under iron deficient conditions <ref name= "spirig"/>. |
'''Class C''' | '''Class C''' | ||
| - | Pili may extend 0.2-3.0 μm from the cell surface and promote cell adhesion and the formation of biofilms. In Gram-positive bacteria class C sortases are used to construct pili, linking together the pilin subunits via isopeptide bonds and only sometimes connects the pilus to the cell wall itself. The general process is conserved, but there is a greater variety in the structure or number of sortases and accessory factors needed | + | Pili may extend 0.2-3.0 μm from the cell surface and promote cell adhesion and the formation of biofilms. In Gram-positive bacteria class C sortases are used to construct pili, linking together the pilin subunits via isopeptide bonds and only sometimes connects the pilus to the cell wall itself. The general process is conserved, but there is a greater variety in the structure or number of sortases and accessory factors needed <ref name= "spirig"/>. The recognition sequence for class C sortases is QVPTG <ref name= "Handbook of Proteolytic Enzymes" />. |
'''Class D''' | '''Class D''' | ||
| - | Class D sortases have so far only been studied in ''B. anthracis'', where it was found to attach BasH and BasI to the cell wall. The class D sortase manages to attach each protein to different structures in the sporulating cell. Deleting the class D sortase reduced the efficiency of sporulation | + | Class D sortases have so far only been studied in ''B. anthracis'', where it was found to attach BasH and BasI to the cell wall. The class D sortase manages to attach each protein to different structures in the sporulating cell. Deleting the class D sortase reduced the efficiency of sporulation <ref name= "spirig"/>. The recognition sequence for class D sortases is LPNTA <ref name= "Handbook of Proteolytic Enzymes" />. |
'''Class E and F''' | '''Class E and F''' | ||
| - | Less is known about class E and F sortases. Class E sortases are believed to be an alternative housekeeping sortase to class A, as the two are never found in the same genome. They recognize the sequence LPXTG. Class F sortases are found in Actinobacteria, but their function is currently unknown | + | Less is known about class E and F sortases. Class E sortases are believed to be an alternative housekeeping sortase to class A, as the two are never found in the same genome. They recognize the sequence LPXTG. Class F sortases are found in Actinobacteria, but their function is currently unknown <ref name= "spirig"/>. |
Revision as of 18:44, 11 December 2015
This page is setup for Brandon to build his senior project for OU CHEM 4923
Sortase System
| |||||||||||
References
1. McCafferty, Dewey G., and Jeffrey A. Melvin. ‘Sortases’. Handbook of Proteolytic Enzymes. N.p.: Elsevier BV, 2013. 2459–2465. PDF.
2. Spirig, T, EM Weiner, and RT Clubb. ‘Sortase Enzymes in Gram-Positive Bacteria’. Molecular microbiology. 5.82 (27 Oct. 2011): n.pag. 4 Nov. 2015.
3. Maresso, Anthony W., Travis J. Chapa, and Olaf Schneewind. ‘Surface Protein IsdC and Sortase B Are Required for Heme-Iron Scavenging of Bacillus Anthracis▿’. 188.23 (29 Sep. 2006): n.pag. 4 Nov. 2015.
4. Theile, Christopher S, et al. ‘Site-Specific N-Terminal Labeling of Proteins Using Sortase-Mediated Reactions’. Nature Protocols 8.9 (29 Aug. 2013): 1800–1807.
5. Mao, H, et al. ‘Sortase-Mediated Protein Ligation: A New Method for Protein Engineering’. Journal of the American Chemical Society. 9.126 (5 Mar. 2004): n.pag. 4 Nov. 2015.
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 McCafferty, Dewey G., and Jeffrey A. Melvin. ‘Sortases’. Handbook of Proteolytic Enzymes. N.p.: Elsevier BV, 2013. 2459–2465. PDF.
- ↑ 2.0 2.1 2.2 2.3 2.4 Spirig, T, EM Weiner, and RT Clubb. ‘Sortase Enzymes in Gram-Positive Bacteria’. Molecular microbiology. 5.82 (27 Oct. 2011): n.pag. 4 Nov. 2015.