Sandbox myosinkinesin
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
'''Head''' | '''Head''' | ||
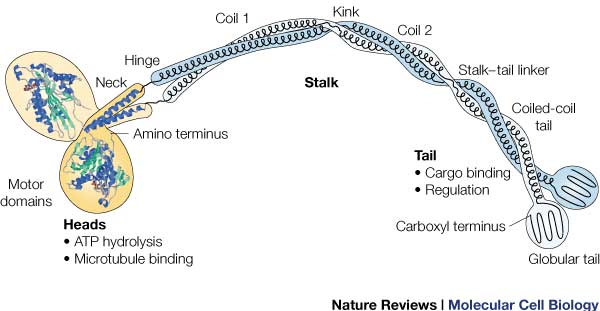
Head: Most conserved domain amongst all kinesin, it consists mainly of alpha helixes. Its tertiary structure usually includes a large cleft from the actin binding site to the ATP binding pocket. The head has the ability to bind microtubule on one site and bind ATP at another. This section undergoes the most conformational change and is responsible for the force that causes kinesin to move along the cytoskeleton. In kinesin, binding of ATP appears to have allosteric control over the binding of kinesin to the tubules, by a twisting of a β- sheet core, showing allosteric control between the subdomains within the protein. | Head: Most conserved domain amongst all kinesin, it consists mainly of alpha helixes. Its tertiary structure usually includes a large cleft from the actin binding site to the ATP binding pocket. The head has the ability to bind microtubule on one site and bind ATP at another. This section undergoes the most conformational change and is responsible for the force that causes kinesin to move along the cytoskeleton. In kinesin, binding of ATP appears to have allosteric control over the binding of kinesin to the tubules, by a twisting of a β- sheet core, showing allosteric control between the subdomains within the protein. | ||
| + | |||
| + | ATPase: It generates this force using an ATPase either coupled with the myosin/kinesin or inherent within the head, the inherent ATPase uses a P-loop, which is a phosphate- binding loop used in adenylate kinase, Ras, and others. Both also create a similar environment for the γ-phosphate, which uses a conserve motif of Ser-Ser-Arg. | ||
| + | |||
| + | In kinesin, the binding of ATP greatly increases the binding of kinesin to the microtubules, to allow it to perform a power stroke. The cleft uses His 93, Pro 17 and Arg 16 to hold ATP. The lack of these loops allows kinesin to react with ATP much faster. | ||
| + | RAS fold: conserved domain among many signaling proteins. A structural motif where a nucleotide molecule is bound to loops at one end of a β-sheet domain. This suggests that myosin and kinesin may have evolved from a common ancestor. | ||
| + | |||
'''Neck''' | '''Neck''' | ||
| + | |||
| + | completely alpha-helical region between the head and the tail on the heavy chain that binds to the light chains. | ||
| + | In kinesin, the neck region of amino acids 330-370 generally form a coiled coil region that is long and flexible. | ||
| + | |||
'''Tail''' | '''Tail''' | ||
| + | : Includes polypeptide binding site, varies the most since it is responsible for dictating the location within the cell that the protein is active. | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 00:29, 16 December 2015
Kinesin
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644