We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox flippases
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | == | + | == Structure == |
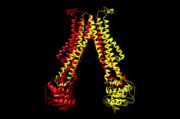
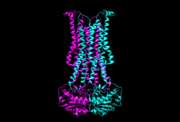
| - | The main three structural features of each monomer are an external helix, a belt of positively charged amino acids, and a binding site for an ATP molecule. | + | Flippase is homodimer which is quartinary strucure. Each monomer is tertiary structure composed with nine alpha-helices and four beta sheets. The secondary structure, helix is composed with amino acids which are primary structure. The main three structural features of each monomer are an external helix, a belt of positively charged amino acids, and a binding site for an ATP molecule. A positively charged belt has four Arginines(R302,R86,R260,R309) which are negative charged. |
== Function == | == Function == | ||
Revision as of 05:08, 16 December 2015
This page is setup for Leah to build her senior project for OU CHEM 4923
Flippases
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Verchère, A., & Menon, A. K. (2015). Structural biology: Lipid gymnastics. Nature, 524(7566), 420-422.