Journal:JBIC:33
From Proteopedia
(Difference between revisions)

| Line 9: | Line 9: | ||
In the field of vaccine development against ''Clostridium difficile'', the NMR assignment and the solution structure of Zmp1, together with its dynamic characterization, can be useful for mapping residues involved in important functions such as eliciting bactericidal antibodies and it will provide insight into improved vaccine antigen design with higher stability and improved epitope presentation. | In the field of vaccine development against ''Clostridium difficile'', the NMR assignment and the solution structure of Zmp1, together with its dynamic characterization, can be useful for mapping residues involved in important functions such as eliciting bactericidal antibodies and it will provide insight into improved vaccine antigen design with higher stability and improved epitope presentation. | ||
| - | *<scene name='71/719223/Cv/5'>Strange bug</scene>. | ||
| - | |||
<scene name='71/719223/Cv/4'>Secondary structure of the zinc-bound Zmp1</scene> ({{Template:ColorKey_Helix}}, | <scene name='71/719223/Cv/4'>Secondary structure of the zinc-bound Zmp1</scene> ({{Template:ColorKey_Helix}}, | ||
{{Template:ColorKey_Strand}}, | {{Template:ColorKey_Strand}}, | ||
| Line 16: | Line 14: | ||
{{Template:ColorKey_Turn}}.) | {{Template:ColorKey_Turn}}.) | ||
| + | <scene name='71/719223/Cv/8'>Zinc coordination site</scene> is shown. The amino acids in the active site are labeled. | ||
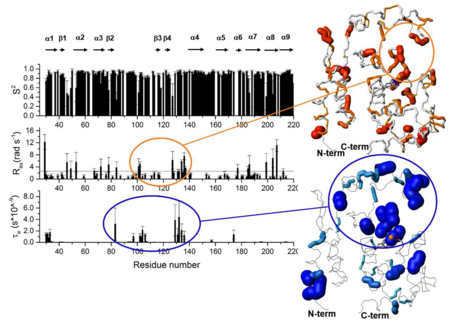
[[Image:JBIC33.png|left|450px|thumb|Parameters characterizing the overall and internal mobility of Zn-bound Zmp1 within the Lipari-Szabo model.]] | [[Image:JBIC33.png|left|450px|thumb|Parameters characterizing the overall and internal mobility of Zn-bound Zmp1 within the Lipari-Szabo model.]] | ||
{{Clear}} | {{Clear}} | ||
Revision as of 12:29, 16 December 2015
| |||||||||||
- ↑ REF
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.