This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Journal:JBIC:33
From Proteopedia
(Difference between revisions)

| Line 5: | Line 5: | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
''Clostridium difficile'' is a Gram-positive spore-forming anaerobic bacterium which emerged in the last decades as a major cause of antibiotic-associated diarrhea worldwide. Tendency of infection to recur and increase in antibiotic-resistant strains pose new challenges to C. difficile treatment. New-generation small molecules, use of pro-biotics, and fecal microbiota transplantation are all emerging options for prevention and cure of C. difficile infection. Beside the development of new antibiotics, virulence targeting factors and vaccines represent promising strategies, which stimulate the research of new potential antigens and targets for drug design. | ''Clostridium difficile'' is a Gram-positive spore-forming anaerobic bacterium which emerged in the last decades as a major cause of antibiotic-associated diarrhea worldwide. Tendency of infection to recur and increase in antibiotic-resistant strains pose new challenges to C. difficile treatment. New-generation small molecules, use of pro-biotics, and fecal microbiota transplantation are all emerging options for prevention and cure of C. difficile infection. Beside the development of new antibiotics, virulence targeting factors and vaccines represent promising strategies, which stimulate the research of new potential antigens and targets for drug design. | ||
| - | ''C. difficile'' releases the zinc-dependent metalloprotease Zmp1 in the extracellular medium. Different possible targets have been identified for this protein, including the human fibrinogen, leading to the hypothesis that it could play multiple roles during bacterial host infection. This protein can indeed contribute to several aspects of C. difficile pathogenesis ranging from the damage of host tissues, through hydrolysis of the components of extracellular matrix, to the release and cell wall attachment of virulence factors. | + | ''C. difficile'' releases the zinc-dependent metalloprotease Zmp1 in the extracellular medium. Different possible targets have been identified for this protein, including the human fibrinogen, leading to the hypothesis that it could play multiple roles during bacterial host infection. This protein can indeed contribute to several aspects of ''C. difficile'' pathogenesis ranging from the damage of host tissues, through hydrolysis of the components of extracellular matrix, to the release and cell wall attachment of virulence factors. |
The structural and dynamic characterization of substrate-free Zmp1 through NMR have revealed that two regions of the protein are affected by local mobility. Such dynamic behavior could play a role in substrate specificity and offers the possibility to investigate and develop specific inhibitors or mutants, that acting on the internal mobility, may impair the activity of ''C. difficile'' Zmp1 contributing to prevent the dissemination of this pathogen. | The structural and dynamic characterization of substrate-free Zmp1 through NMR have revealed that two regions of the protein are affected by local mobility. Such dynamic behavior could play a role in substrate specificity and offers the possibility to investigate and develop specific inhibitors or mutants, that acting on the internal mobility, may impair the activity of ''C. difficile'' Zmp1 contributing to prevent the dissemination of this pathogen. | ||
In the field of vaccine development against ''Clostridium difficile'', the NMR assignment and the solution structure of Zmp1, together with its dynamic characterization, can be useful for mapping residues involved in important functions such as eliciting bactericidal antibodies and it will provide insight into improved vaccine antigen design with higher stability and improved epitope presentation. | In the field of vaccine development against ''Clostridium difficile'', the NMR assignment and the solution structure of Zmp1, together with its dynamic characterization, can be useful for mapping residues involved in important functions such as eliciting bactericidal antibodies and it will provide insight into improved vaccine antigen design with higher stability and improved epitope presentation. | ||
| Line 13: | Line 13: | ||
{{Template:ColorKey_Loop}}, | {{Template:ColorKey_Loop}}, | ||
{{Template:ColorKey_Turn}}.) | {{Template:ColorKey_Turn}}.) | ||
| + | |||
| + | *<scene name='71/719223/Cv/9'>Click here to see animation of Zn-bound Zmp1 internal mobility</scene>. | ||
<scene name='71/719223/Cv/8'>Zinc coordination site</scene> is shown. The amino acids in the active site are labeled. | <scene name='71/719223/Cv/8'>Zinc coordination site</scene> is shown. The amino acids in the active site are labeled. | ||
| Line 25: | Line 27: | ||
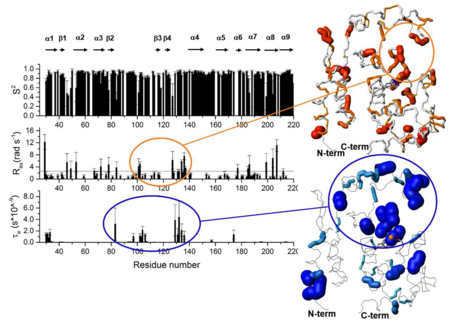
scales (ms to t<sub>s</sub> ), characteristic of exchange processes (R<sub>ex</sub>), may also contribute to | scales (ms to t<sub>s</sub> ), characteristic of exchange processes (R<sub>ex</sub>), may also contribute to | ||
transverse relaxation through the fluctuation of the chemical environment of a nucleus. | transverse relaxation through the fluctuation of the chemical environment of a nucleus. | ||
| - | The line reported in the graph of | + | The line reported in the graph of R<sub>ex</sub> represents the average value. The secondary |
structure elements are reported at the top. Residues which experience local mobility (R<sub>ex</sub> | structure elements are reported at the top. Residues which experience local mobility (R<sub>ex</sub> | ||
or t<sub>e</sub>) are also mapped into the Zmp1 structure in orange and blue respectively. The | or t<sub>e</sub>) are also mapped into the Zmp1 structure in orange and blue respectively. The | ||
| Line 33: | Line 35: | ||
[[Image:JBIC33.png|left|450px|thumb|]] | [[Image:JBIC33.png|left|450px|thumb|]] | ||
{{Clear}} | {{Clear}} | ||
| - | + | ||
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Revision as of 12:53, 16 December 2015
| |||||||||||
- ↑ REF
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.