Sandbox myosinkinesin
From Proteopedia
(Difference between revisions)
| Line 29: | Line 29: | ||
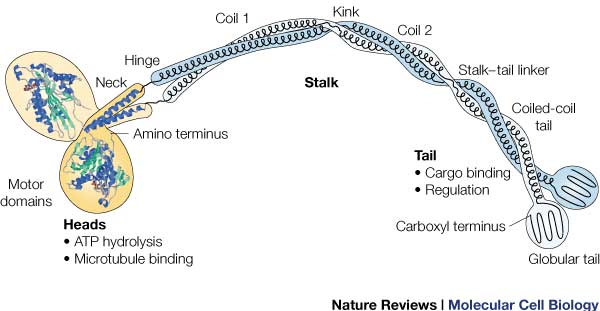
ATPase: It generates this force using an ATPase either coupled with the myosin/kinesin or inherent within the head, the inherent ATPase uses a P-loop, which is a phosphate- binding loop used in adenylate kinase, Ras, and others. Both also create a similar environment for the γ-phosphate, which uses a conserve motif of Ser-Ser-Arg.<ref name= "RAS">doi: 10.1038/380550a0</ref> | ATPase: It generates this force using an ATPase either coupled with the myosin/kinesin or inherent within the head, the inherent ATPase uses a P-loop, which is a phosphate- binding loop used in adenylate kinase, Ras, and others. Both also create a similar environment for the γ-phosphate, which uses a conserve motif of Ser-Ser-Arg.<ref name= "RAS">doi: 10.1038/380550a0</ref> | ||
| - | In kinesin, the binding of ATP greatly increases the binding of kinesin to the microtubules, to allow it to perform a power stroke | + | In kinesin, the binding of ATP greatly increases the binding of kinesin to the microtubules, to allow it to perform a power stroke. |
RAS fold: conserved domain among many signaling proteins. A structural motif where a nucleotide molecule is bound to loops at one end of a β-sheet domain. This suggests that myosin and kinesin may have evolved from a common ancestor.<ref name= "RAS"/> | RAS fold: conserved domain among many signaling proteins. A structural motif where a nucleotide molecule is bound to loops at one end of a β-sheet domain. This suggests that myosin and kinesin may have evolved from a common ancestor.<ref name= "RAS"/> | ||
Revision as of 18:22, 17 December 2015
Kinesin
| |||||||||||
References
- ↑ doi: https://dx.doi.org/10.2210/rcsb_pdb/mom_2005_4
- ↑ Hackney, David. "Kinesin-1 Structure." Duke. Duke, 25 Jan. 2005. Web. 17 Dec. 2015. <https://labs.cellbio.duke.edu/kinesin/KinesinStructure.html>.
- ↑ Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993 Jul 2;261(5117):50-8. PMID:8316857
- ↑ Krukau A, Knecht V, Lipowsky R. Allosteric control of kinesin's motor domain by tubulin: a molecular dynamics study. Phys Chem Chem Phys. 2014 Apr 7;16(13):6189-98. doi: 10.1039/c3cp53367k. PMID:24561904 doi:http://dx.doi.org/10.1039/c3cp53367k
- ↑ 5.0 5.1 Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996 Apr 11;380(6574):550-5. PMID:8606779 doi:http://dx.doi.org/10.1038/380550a0
- ↑ 6.0 6.1 Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999 Dec 16;402(6763):778-84. PMID:10617199 doi:http://dx.doi.org/10.1038/45483
- ↑ Song YH, Marx A, Muller J, Woehlke G, Schliwa M, Krebs A, Hoenger A, Mandelkow E. Structure of a fast kinesin: implications for ATPase mechanism and interactions with microtubules. EMBO J. 2001 Nov 15;20(22):6213-25. PMID:11707393 doi:10.1093/emboj/20.22.6213