We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox T4SS
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | This page is setup for Jian to build her senior project for OU CHEM 4923 | + | This page is setup for Jian to build her senior project for OU CHEM 4923 |
==FUNCTIONS OF TYPE 4 SECRETION SYSTEMS== | ==FUNCTIONS OF TYPE 4 SECRETION SYSTEMS== | ||
<StructureSection load='1gki' size='340' side='right' caption='TwrB hexamer a homologue for the VirD4 from ''E. coli''' scene=''> | <StructureSection load='1gki' size='340' side='right' caption='TwrB hexamer a homologue for the VirD4 from ''E. coli''' scene=''> | ||
| - | Bacterial type IV secretions systems move DNA and proteins out of a cell. Usually transfer of this DNA/proteins requires cell-to-cell contact, however one type of T4SS releases DNA from the extracellular milieu<ref | + | Bacterial type IV secretions systems move DNA and proteins out of a cell. Usually transfer of this DNA/proteins requires cell-to-cell contact, however one type of T4SS releases DNA from the extracellular milieu. <ref name="one">doi:10.1111/j.1462-5822.2010.01499.x</ref> |
| - | + | ||
| - | + | ||
==GENERAL STRUCTURE FEATURES OF TYPE 4 SECRETION SYSTEMS== | ==GENERAL STRUCTURE FEATURES OF TYPE 4 SECRETION SYSTEMS== | ||
| - | With different functions comes many variations on the structure of T4SS, however the most well understood set of structures were studied through the Gram-negative bacteria ''A. tumefaciens''. It is called the VirB/VirD4 system. | + | With different functions comes many variations on the structure of T4SS, however the most well understood set of structures were studied through the Gram-negative bacteria ''A. tumefaciens''. It is called the VirB/VirD4 system<ref name="two">doi:10.1016/j.bbamcr.2013.12.019</ref>. |
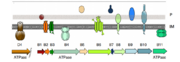
VirB is made up of 11 different VirB proteins systhesized from the ''virB'' operon. Most T4SS have homologes of these 12 proteins (11 from VirB and 1 from VirD) | VirB is made up of 11 different VirB proteins systhesized from the ''virB'' operon. Most T4SS have homologes of these 12 proteins (11 from VirB and 1 from VirD) | ||
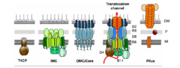
| - | + | VirB7, VirB7 and VirB10 initiate the assembly of the core of the T4SS. The complex forms spontaneously with out the input of external energy like ATP. [[Image:Janeyuan1.2.png|thumb|Overview of formation of T4SS<ref name="one" />]] | |
| - | [[Image:Janeyuan1.1.png|thumb|Overview of T4SS structures<ref | + | [[Image:Janeyuan1.1.png|thumb|Overview of T4SS structures<ref name="one" />]] |
----- | ----- | ||
| - | == Function == | ||
| - | |||
| - | == Disease == | ||
| - | |||
| - | == Relevance == | ||
| - | |||
| - | == Structural highlights == | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 19:03, 17 December 2015
This page is setup for Jian to build her senior project for OU CHEM 4923
FUNCTIONS OF TYPE 4 SECRETION SYSTEMS
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Wallden K, Rivera-Calzada A, Waksman G. Type IV secretion systems: versatility and diversity in function. Cell Microbiol. 2010 Sep 1;12(9):1203-12. doi: 10.1111/j.1462-5822.2010.01499.x. , Epub 2010 Jul 16. PMID:20642798 doi:http://dx.doi.org/10.1111/j.1462-5822.2010.01499.x
- ↑ Christie PJ, Whitaker N, Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014 Aug;1843(8):1578-91. doi:, 10.1016/j.bbamcr.2013.12.019. Epub 2014 Jan 2. PMID:24389247 doi:http://dx.doi.org/10.1016/j.bbamcr.2013.12.019