Adrenodoxin reductase
From Proteopedia
(Difference between revisions)
| Line 47: | Line 47: | ||
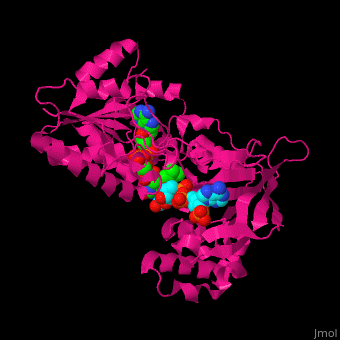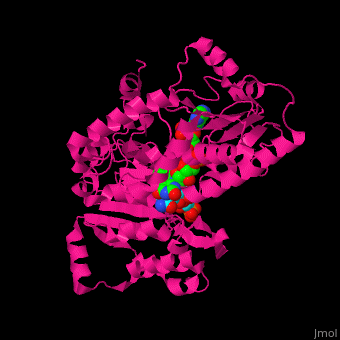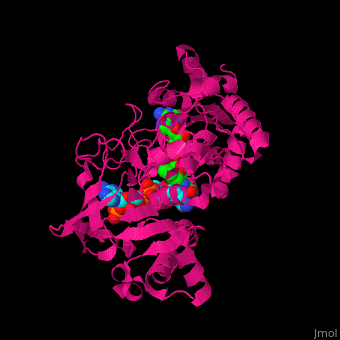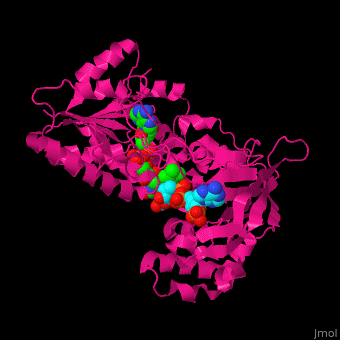
<scene name='70/702915/Cv/9'>FAD binding site in AR</scene> (water molecules shown as red spheres) | <scene name='70/702915/Cv/9'>FAD binding site in AR</scene> (water molecules shown as red spheres) | ||
| - | <scene name='70/702915/Cv/10'>NADP binding site in AR</scene> (water molecules shown as red spheres) | + | <scene name='70/702915/Cv/10'>NADP binding site in AR</scene> (water molecules shown as red spheres) (PDB entry [[1e1k]])<ref>PMID:10998235</ref> |
| - | <scene name='70/702915/Fad_rossmann_fold/1'>Rossmann fold of FAD</scene> | + | <scene name='70/702915/Fad_rossmann_fold/1'>Rossmann fold of FAD</scene> |
{{clear}} | {{clear}} | ||
</StructureSection> | </StructureSection> | ||
Revision as of 10:08, 11 January 2016
| |||||||||||
3D Structures of Adrenodoxin reductase
Updated on 11-January-2016
1e1k, 1e1l, 1e1m – bADR+FAD+NADP – bovine
1e1n, 1cjc – bADR+FAD
1e6e – bADR + adrenodoxin
References
- ↑ 1.0 1.1 Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol. 1992 Dec;43(8):779-804. doi:, 10.1016/0960-0760(92)90307-5. PMID:22217824 doi:http://dx.doi.org/10.1016/0960-0760(92)90307-5
- ↑ Ziegler GA, Schulz GE. Crystal structures of adrenodoxin reductase in complex with NADP+ and NADPH suggesting a mechanism for the electron transfer of an enzyme family. Biochemistry. 2000 Sep 12;39(36):10986-95. PMID:10998235
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Israel Hanukoglu, Michal Harel, Joel L. Sussman