Apurinic-Apyrimidinic Endonuclease
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load='2o3c' size='350' side='right' caption=' | + | <StructureSection load='2o3c' size='350' side='right' caption='Zebrafish APE1 trimer interacting with Pb+2 ions. [[2o3c]]' scene=''> |
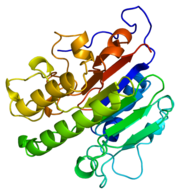
[[Image:thumbnail.png|thumb|left|Figure 1.Ribbon Structure of APE1. Antiparallel beta sheets line the interior of the enzyme, while alpha helices line the exterior.]] | [[Image:thumbnail.png|thumb|left|Figure 1.Ribbon Structure of APE1. Antiparallel beta sheets line the interior of the enzyme, while alpha helices line the exterior.]] | ||
| Line 14: | Line 14: | ||
==Substrate Binding and Enzymatic Mechanism== | ==Substrate Binding and Enzymatic Mechanism== | ||
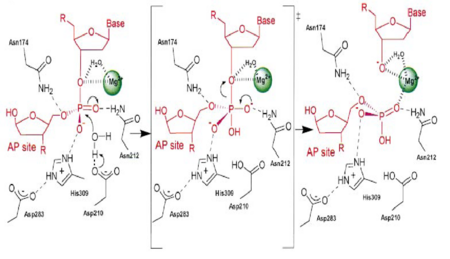
| - | [[Image:APE1mech.png|thumb|Figure 3. APE1 enzymatic mechanism of phosphodiester bond cleavage of abasic DNA<ref name="Mol"/>.]] | + | [[Image:APE1mech.png|left|450px|thumb|Figure 3. APE1 enzymatic mechanism of phosphodiester bond cleavage of abasic DNA<ref name="Mol"/>.]] |
| - | + | {{Clear}} | |
APE1 has numerous substrates in which in binds to ands acts on enzymatically, with abasic DNA being the most well known substrate for APE1. APE1 binds on the 5' side of the abasic DNA site with the amino acid residues Arg73, Ala74 and Lys78 interacting with three consecutive phosphate molecules on the DNA strand, while Tyr128 and Gly127 simultaneously span the minor grove causing the groove to widen a distance of ~2 Angstroms<ref name="Mol"/>.The DNA is anchored and the abasic DNA is stabalized by APE1. APE1 then inserts Met270 into the minor groove causing the APE1/DNA structure to become extremely kinked<ref name="Mol"/>.This kinking of the DNA strand is followed by an Arg177 insertion to the major groove donating a hydrogen bond to the 3' phosphate located at the abasic site<ref name="Mol"/>.This is followed by a nucleophilic attack of an aspartate-activated hydroxyl group forming a transition state that is stabalized by a Mg2+ ion (Figure 3) which is followed by cleavage of the phosphodiester bond on the DNA strand<ref name="Mol"/>.This is followed by the sequential events of residue Trp280 contacting a DNA phosphate and residues Asn222, Asn226 and Asn229 contacting the phosphates 3' of the abasic site, which is followed by Asn174 and Asn212 forming hydrogen bonds with an oxygen molecules 5' to the abasic site<ref name="Mol"/>. | APE1 has numerous substrates in which in binds to ands acts on enzymatically, with abasic DNA being the most well known substrate for APE1. APE1 binds on the 5' side of the abasic DNA site with the amino acid residues Arg73, Ala74 and Lys78 interacting with three consecutive phosphate molecules on the DNA strand, while Tyr128 and Gly127 simultaneously span the minor grove causing the groove to widen a distance of ~2 Angstroms<ref name="Mol"/>.The DNA is anchored and the abasic DNA is stabalized by APE1. APE1 then inserts Met270 into the minor groove causing the APE1/DNA structure to become extremely kinked<ref name="Mol"/>.This kinking of the DNA strand is followed by an Arg177 insertion to the major groove donating a hydrogen bond to the 3' phosphate located at the abasic site<ref name="Mol"/>.This is followed by a nucleophilic attack of an aspartate-activated hydroxyl group forming a transition state that is stabalized by a Mg2+ ion (Figure 3) which is followed by cleavage of the phosphodiester bond on the DNA strand<ref name="Mol"/>.This is followed by the sequential events of residue Trp280 contacting a DNA phosphate and residues Asn222, Asn226 and Asn229 contacting the phosphates 3' of the abasic site, which is followed by Asn174 and Asn212 forming hydrogen bonds with an oxygen molecules 5' to the abasic site<ref name="Mol"/>. | ||
==The nuclease domain== | ==The nuclease domain== | ||
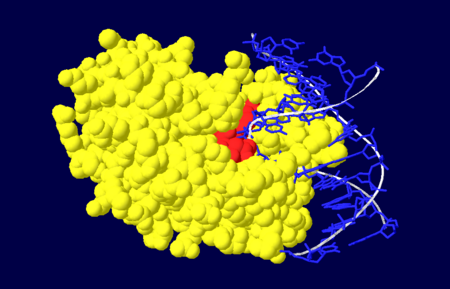
| - | [[Image:NUDOM.png|thumb|Figure 4. Nuclease domain (yellow) of APE1 bound to DNA (blue), with the active site amino acid residues labelled in red. <ref name="Mol"/>.]] | + | [[Image:NUDOM.png|left|450px|thumb|Figure 4. Nuclease domain (yellow) of APE1 bound to DNA (blue), with the active site amino acid residues labelled in red. <ref name="Mol"/>.]] |
| + | {{Clear}} | ||
The nuclease domain of APE1 involves amino acid residues 128-318<ref name="Masood"/>. The nuclease activity of APE1 is due to its positively charged active site and its ability to bind to both the abasic and opposing DNA strand which leads to the displacement of the bound DNA glycosylase enzyme in the base excision repair pathway, allowing the DNA strand to be repaired<ref name="Mol"/>. | The nuclease domain of APE1 involves amino acid residues 128-318<ref name="Masood"/>. The nuclease activity of APE1 is due to its positively charged active site and its ability to bind to both the abasic and opposing DNA strand which leads to the displacement of the bound DNA glycosylase enzyme in the base excision repair pathway, allowing the DNA strand to be repaired<ref name="Mol"/>. | ||
Revision as of 11:08, 12 January 2016
| |||||||||||
3D structures of Apurinic/Apyrimidinic Endonuclease
Updated on 12-January-2016
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879
- ↑ 2.0 2.1 Barnes, T., et al. 2009. Identification of Apurinic/apyrimidinic endoribonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA in vitro. Nucleic Acid Research 37: 3946-3958
- ↑ 3.0 3.1 3.2 Ando, K. et al. 2008. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kB and AP-1, and activation of their DNA binding activity. Nucleic Acid research 36(13): 4327-4336.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 4.9 Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
Proteopedia Page Contributors and Editors (what is this?)
Mark Barnes, Michal Harel, Alexander Berchansky, David Canner, Jaime Prilusky, Andrea Gorrell, Eran Hodis, Joel L. Sussman