, also known as BACE (β-site of Amyloid precursor protein Cleaving Enzyme) and Memapsin 2, is an aspartyl protease found in humans. β-Secretase plays an important role in the development of Alzheimer's Disease. This has made β-secretase a therapeutic target for pharmacological intervention. See also Beta-secretase 1.
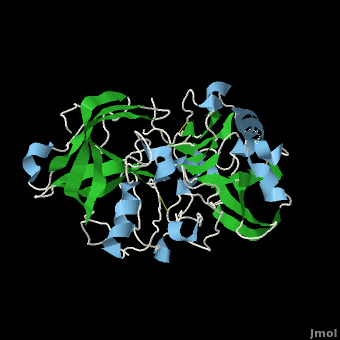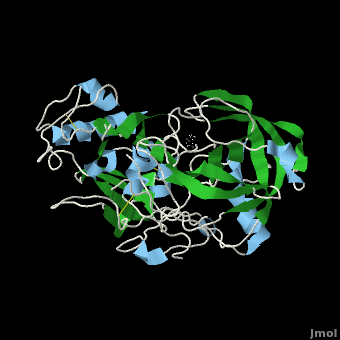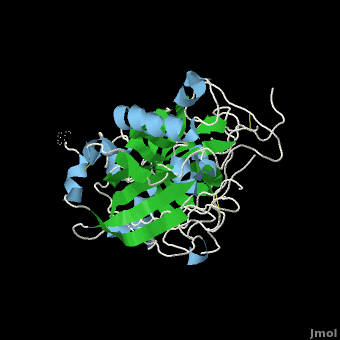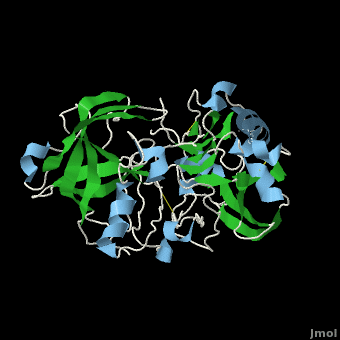
Structure of Beta Secretase
β-Secretase is a transmembrane protein, with its active site existing in the extracellular domain of the protein. Due to its nature as an aspartyl protease, β-secretase's active site is made up of . The R groups of both aspartates coordinate a single water molecule between the two of them, allowing for a nucleophilic attack to occur on the carbonyls.
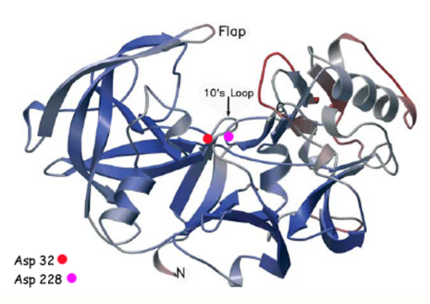
There are two other important features to β-secretase. One is the . The flap is made up of residues 67 through 77. While the active site remains inactive, the flap stays in its open conformation. However, the flap is stabilized while closed over its substrate or some other inhibitor.
The other important feature is the . The 10s loop is located in the S3 pocket of β-secretase, right between two β strands. When the 10s loop takes an open conformation, it allows for greater binding between the substrate and the S3 pocket. The 10s loop also contains within it a with which the substrate can form a hydrogen bond, allowing for further stabilization of the 10s loop, as well as the overall β-secretase-substrate interaction.
The three parts come together to form a sort of binding pocket for β-secretase's substrates or inhibitors. Binding to the active site activates the flap to close and initiates binding by the 10s loop, all to help stabilize the structure.
Alzheimer's Disease
Alzheimer's Disease is a neurodegenerative disease, and one of the most common forms of dementia. Early symptoms include things like loss of short-term memory and impairment of some physical movements. As the disease progresses, memory impairment worsens, muscle deteriorates, and the patient eventually dies. Patients of the later stages of Alzheimer's usually need to be taken care of as they are not able to take care of themselves at that point.
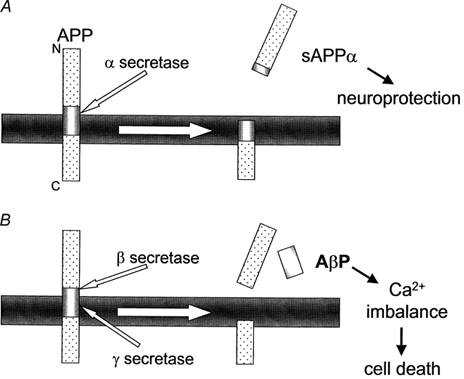
Alzheimer's disease occurs by plaques in the walls of vessels and extracellular parenchyma in the brain that lead to the death of neurons. The plaques themselves are formed from the buildup of amyloid beta (Aβ). Aβ is a fragment of about 43 amino acids, and is produced by the cleavage of amyloid precursor protein (APP), an integral membrane protein found in the synapses of neurons. The cleavage of APP itself can go down one of two pathways:
A. In the first pathway, α-secretase cleaves APP somewhere within the Aβ region. This creates a fragment known as sAPPα. This fragment is beneficial to neurons as it helps to protect them. γ-Secretase can also follow up and cleave at its target, forming a fragment known as p3, whose function is unknown.
B. In the second pathway, β-secretase cleaves APP at the N-terminus of Aβ, creating a fragment of sAPPβ. Then γ-secretase cleaves APP at the C-terminus of Aβ, which exists along the transmembrane domain of APP. At this point, Aβ is released and allowed to accumulate with other fragments, forming plaques.
β-Secretase is able to cleave Aβ at its N-terminus due to the nucleophilic attack that occurs upon the the active site of β-secretase. This β-site region which β-secretase binds to is made up of methionine-aspartate-alanine (MDA). The aspartates on β-secretase bind to the aspartate of the β-site on APP (Asp672). This reaction only occurs under acidic pH conditions. After the water molecule is coordinated between the carbonyls of the aspartates and the amine group and R group hydroxyl of the aspartate on Aβ, the two are able to react, forcing the N-terminus to break its bond with sAPPβ.
β-Secretase has a specific binding region, where it only binds to the MDA sequence on APP. Mutations and/or radical insertions in the β-site keeps β-secretase from binding to APP. Additionally, shortening of the 10s loop also causes β-secretase to lose its binding ability. This either comes from Gly11 being too far to bond with the substrate, or just the removal of Gly11 as part of the loop. Without the loop, β-secretase is unable to stabilize its interaction with APP and is unable to cleave it.
Numerous studies performed on mice have shown that β-secretase -/- knockouts produce no Aβ. Additionally, the mice were examined for behavioral defects and were reported normal. This information encourages the idea that β-secretase may be unnecessary. However, there have been recent studies done that show that lack of β-secretase and Aβ can actually lead to some behavioral defects, leading researchers to look more into this cause.
Inhibition of Beta Secretase
Due to β-secretase's function in the production of Aβ, it has become a very popular target for therapeutic drugs. An example of one such developed drug is (), which comes from Astex Technology.
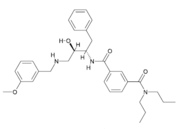
Fig. 1. Structure of OM99-2.
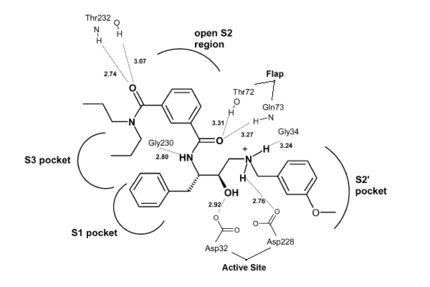
Once the inhibitor moves into place, its positively charged amine group and its hydroxyl group start to interact with β-secretase's active site. The nucelophilic attack on the aspartate's carbonyls binds OM99-2 to β-secretase. As OM99-2 becomes situated within β-secretase's binding pocket, the flap closes upon OM99-2. The flap's residues Thr72 and Gln73 bind with one of OM99-2's carbonyl groups. The 10s loop remains open to allow OM99-2 to interact with the S3 pocket. Gly11 also forms a hydgrogen bond using its carbonyl with the amino terminus of OM99-2. At this point OM99-2 is locked securely within .
OM99-2 remains stabilized in the pocket by binding to the other pockets of β-secretase (S1, S2, S2', and S3). OM99-2 complexed with β-secretase shows the hydrophobic nature of the S1 and S3 pockets. Additionally, OM99-2 is able to interact with other residues in β-secretase (in this case Gly34 and Thr232), though this is not the case for all inhibitors.
Numerous tests have been conducted with OM99-2 as an inhibitor, the results have been mainly positive. However, there have occurred a substantial number of times results where the flap only partially closes and the 10s loop closes off the S3 pocket. While the inhibitor works well when it is stabilized within β-secretase, the fact that it does not always stabilize keeps the inhibitor from being consumed by humans. Additionally, β-secretase must be soaked in OM99-2 to assure that it binds to the inhibitor.
The apo structure of β-secretase was obtained by refolding recombinant protein developed with E. coli. The β-secretase/OM99-2 crystal structure was obtained by soaking β-secretase with OM99-2 and stabilizing the entire interaction with iodide ions. Mass spectrometry and X-ray crytstallography were used to analyze the obtained structures.
Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
is a transmembrane aspartic protease that cleaves the β-amyloid precursor protein en route to generation of the amyloid β-peptide (Aβ) that is believed to be the principal component of the amyloid plaques seen in the brains of Alzheimer’s disease. It is thus a prime target for the development of inhibitors that may serve as drugs in the treatment and/or prevention of Alzheimer’s disease. The protease domain has a conserved aspartic protease fold, with the (colored cyan and yellow, respectively). The hairpin loop in the N-terminal lobe that is known as the , as is in general the case for the aspartic protease family. Determination of the crystal structures of both apo and complexed BACE1, structural analysis of all crystal structures of BACE1 deposited in the PDB, and MD simulations of and BACE1 were used to study conformational changes in the active-site region of the enzyme (the first monomer in magenta with a red flap, the second one in green with an orange flap). Four new crystal structures were determined: that of a P6122 (3tpr, BACE1 in magenta and in cyan), those of a (3tpp, BACE1 in indigo and in green) and of an (3tpj, in orange), both belonging to space group C2221, and that of a (3tpl, in blueviolet). P6122 (3tpr, BSIIV/BACE1) were obtained by , while those of the complex belonging to space group C2221 were obtained by (3tpp, BSIIV/BACE1). The two apo structures belonging to different space groups were obtained by cocrystallization in the presence of another low affinity inhibitor, using two different precipitants; in both cases the inhibitor was not seen in the crystals thus obtained. It was found that the , as do the , particularly in the . This led to a systematic analysis of all the BACE1 crystal structures deposited in the PDB (both apo structures and complexes), and to perform MD simulations on the apo enzyme, so as to investigate the underlying causes for the various conformations adopted by the flap. In particular, an attempt was made to clarify whether they might be a consequence of crystal packing in different space groups, or might indicate that the active site can assume multiple low-energy conformations. The RMSFs of main-chain atoms, calculated based on all the BACE1 crystal structures deposited in the PDB, show that the . The , reveals that the (e.g. 1w50, colored deepskyblue) with two exceptions; not surprisingly, the situation with respect to the structures of the complexes is more complex. (e.g. 3tpp, BSIIV/BACE1), except in a few cases in which the position and orientation of the inhibitors sterically preclude its closing. It is of interest that (3tpr, BSIIV/BACE1); but a direct H-bond between Q73 and BSIIV locks the flap in a closed conformation in the co-crystallized structure (3tpp, BSIIV/BACE1). It may thus be concluded that formation of a direct H-bond between a main-chain atom of a flap residue and the bound inhibitor is a prerequisite for the flap to assume a closed conformation in a complex. Interestingly, P6122 except one, the flap is in an open conformation. Our soaked crystals of the BSIIV/BACE1 complex also belong to space group P6122. Analysis of the crystal packing pattern reveals that in this space group the flap is at a packing interface. MD simulations were carried out to check if the observed packing would influence the conformation of the flap. In the starting model for MD that includes a symmetry-related copy of BACE1, both the degree of opening and the time that the flap dwells in the open state are much larger than in the trajectories generated for the monomer. Thus, the packing pattern in crystals belonging to space group P6122 hinders the conformational change in the flap, which may be the main reason why in our BSIIV/BACE1 complex (3tpr) belonging to space group P6122 the C2221 (3tpp, BSIIV/BACE1). Thus a systematic study of the conformational flexibility of BACE1 may not only contribute to structure-based drug design of BACE1 inhibitors, but also produce a better understanding of the mechanistic events associated with binding of substrates and inhibitors to the enzyme.