Sandbox Reserved 1121
From Proteopedia
| Line 29: | Line 29: | ||
=== Phosphocholine-binding site === | === Phosphocholine-binding site === | ||
| - | PC stands for phosphocholine. It is a phospholipid in cell membranes and a plasma lipoprotein <ref name=" | + | PC stands for phosphocholine. It is a phospholipid in cell membranes and a plasma lipoprotein <ref name="Thompson"/>. The PC-binding site is a hydrophobic pocket constituted by the residues Leu64, Phe66, Thr76 and the two Ca<sup>2+</sup> <ref name="agrawal"/>. |
The choline function of PC interacts with the two key residues Phe66 and Glu81, therefore PC lies inside the PC-binding site <ref name="kumar"/> <ref name="agrawal"/>. | The choline function of PC interacts with the two key residues Phe66 and Glu81, therefore PC lies inside the PC-binding site <ref name="kumar"/> <ref name="agrawal"/>. | ||
The PC-binding site is next to the Ca<sup>2+</sup>-binding sites on the same face of the CRP protein. The phosphate groupe of PC interacts by coordination with the two Ca<sup>2+</sup> <ref name="agrawal"/>. | The PC-binding site is next to the Ca<sup>2+</sup>-binding sites on the same face of the CRP protein. The phosphate groupe of PC interacts by coordination with the two Ca<sup>2+</sup> <ref name="agrawal"/>. | ||
| Line 40: | Line 40: | ||
== Biomedical interest == | == Biomedical interest == | ||
| - | Healthy humans have a CRP rate which is generally about 1 μg/mL<ref name="kumar"/>. CRP is secreted by the liver into the blood circulation<ref name = "Alex"/>. CRP level is 1000 times higher in a cytokine-mediated response due to tissue injury, infection and inflammation. Therefore the CRP rate in serum is common use to detect the activity of a disease<ref name=" | + | Healthy humans have a CRP rate which is generally about 1 μg/mL<ref name="kumar"/>. CRP is secreted by the liver into the blood circulation<ref name = "Alex"/>. CRP level is 1000 times higher in a cytokine-mediated response due to tissue injury, infection and inflammation. Therefore the CRP rate in serum is common use to detect the activity of a disease<ref name="Thompson" />. CRP can be defined as a target for the development of cardioprotection and neuroprotection<ref name="kumar"/>. |
== Structural highlights == | == Structural highlights == | ||
Revision as of 09:23, 28 January 2016
|
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Contents |
Human C-reactive protein complexed with phosphocholine
Human C-Reactive Protein (CRP) is an acute phase protein belonging to the highly conserved pentraxin protein family, like as its homologue, the Serum Amyloid P component (SAP) [1]. Although it is a normal serum protein, its circulating concentration raises rapidly and extensively in a cytokine-mediated in response to an infection, an inflammation or a tissue injury. In this way, serum CRP rate is empirically dose to detect many human disease [1]. CRP was named this way because it was found that the protein could precipitate the "C" polysaccharide derived from Streptococcus pneumoniae cell wall [2][3].
Structure
CRP structure
The crystal structure of CRP was determined by using SAP as the search model [1]. The structure of CRP has been determined by X-ray crystallograph at 3 Å resolution.
Primary structure: The C-reactive protein is a homopentamer in which each subunit is a 25 kDa protein consisting of 224 residues [4]. Ser53, His95, Cys97, Asp112, Gly113, Gly136, Gly154, Val165, Leu166, Ile171, and Gly196 are the highly conserved residues in the primary sequence of CRP [3].
Secondary structure: The secondary structure is formed of one and two antiparallel [4]. The predominant structure is β-sheet [5] but short helical regions can be noticed for the residues 43 and 185 [3]. The residues Glu197 and Lys123 of CRP form an intermolecular ion pair [1].
Tertiary and quaternary structure: Like SAP, the protein consists of five protomers, uncovalently bonded and nonglycosylated that are arranged symmetrically around a central pore [6] [7]. The diameter of the CRP pentamer is 102 Å, the inner pore diameter is 30 Å and the diameter of a subunit is 36 Å [8]. Thereafter, the ligand-binding face of the molecule will be called B and the opposite face will be called A. The B face binds two Ca2+ ions and a phosphocholine per subunit. The A face, recognizable by the presence of an α-helix and a deep cavity, can interact with C1q and Fc receptors [7].
Ca2+-binding site
CRP is a calcium dependent structure. In fact, Ca2+ is required for PC binding, and more precisely for the formation of the PC-binding site thanks to structural rearrangements. The protection against denaturation and proteolysis is also performed through Ca2+-binding. In the absence of Ca2+, hCRP is cleaved between Asn145 and Phe146 by nagarse protease, and between Phe146 and Glu147 by pronase. Asp60, Asn61, Glu138, Asp140 and the main-chain carbonyl of Gln139 residues allow the first calcium ion binding, and the second is performed through Glu138, Asp140, Glu147 and Gln150 [9]. The two Ca2+-binding sites are overlapping in a loop. In the absence of Ca2+, the loop changes conformation and releases the proteolysis site. Therefore Ca2+ protects CRP form proteolytic cleavage [8].
Phosphocholine-binding site
PC stands for phosphocholine. It is a phospholipid in cell membranes and a plasma lipoprotein [1]. The PC-binding site is a hydrophobic pocket constituted by the residues Leu64, Phe66, Thr76 and the two Ca2+ [8]. The choline function of PC interacts with the two key residues Phe66 and Glu81, therefore PC lies inside the PC-binding site [3] [8]. The PC-binding site is next to the Ca2+-binding sites on the same face of the CRP protein. The phosphate groupe of PC interacts by coordination with the two Ca2+ [8]. The affinity of CRP for PC increases with the concentration of PC. A surface containing a high density of PC, such as C-polycaccharide, is therefore propitious to the CRP-binding [7]. CRP can also bind chromatin, histones, small nuclear ribonucleoproteins nuclear envelop proteins and nucleosomes Ca2+-dependently [8].
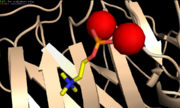
Function
CRP binds to PC located on the surface of bacteria that infected the organism. The resulting immune response is the phygocytosis of PC-expressing bacteria [8]. The CRP is therefore part of the acute phase response which is a rapid concentration variation of plasma proteins [11].
Biomedical interest
Healthy humans have a CRP rate which is generally about 1 μg/mL[3]. CRP is secreted by the liver into the blood circulation[11]. CRP level is 1000 times higher in a cytokine-mediated response due to tissue injury, infection and inflammation. Therefore the CRP rate in serum is common use to detect the activity of a disease[1]. CRP can be defined as a target for the development of cardioprotection and neuroprotection[3].
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure. 1999 Feb 15;7(2):169-77. PMID:10368284
- ↑ Black S, Kushner I, Samols D. C-reactive Protein. J Biol Chem. 2004 Nov 19;279(47):48487-90. Epub 2004 Aug 26. PMID:15337754 doi:http://dx.doi.org/10.1074/jbc.R400025200
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Kumar SV, Ravunny RK, Chakraborty C. Conserved domains, conserved residues, and surface cavities of C-reactive protein (CRP). Appl Biochem Biotechnol. 2011 Sep;165(2):497-505. doi: 10.1007/s12010-011-9270-7., Epub 2011 May 4. PMID:21541851 doi:http://dx.doi.org/10.1007/s12010-011-9270-7
- ↑ 4.0 4.1 UniProtKB - P02741 (CRP_HUMAN)
- ↑ Dong A, Caughey B, Caughey WS, Bhat KS, Coe JE. Secondary structure of the pentraxin female protein in water determined by infrared spectroscopy: effects of calcium and phosphorylcholine. Biochemistry. 1992 Oct 6;31(39):9364-70. PMID:1382589
- ↑ Volanakis JE. Human C-reactive protein: expression, structure, and function. Mol Immunol. 2001 Aug;38(2-3):189-97. PMID:11532280
- ↑ 7.0 7.1 7.2 Du Clos TW, Mold C. C-reactive protein: an activator of innate immunity and a modulator of adaptive immunity. Immunol Res. 2004;30(3):261-77. PMID:15531769 doi:http://dx.doi.org/10.1385/IR:30:3:261
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 Agrawal A, Singh PP, Bottazzi B, Garlanda C, Mantovani A. Pattern recognition by pentraxins. Adv Exp Med Biol. 2009;653:98-116. PMID:19799114
- ↑ Ramadan MA, Shrive AK, Holden D, Myles DA, Volanakis JE, DeLucas LJ, Greenhough TJ. The three-dimensional structure of calcium-depleted human C-reactive protein from perfectly twinned crystals. Acta Crystallogr D Biol Crystallogr. 2002 Jun;58(Pt 6 Pt 2):992-1001. Epub, 2002 May 29. PMID:12037301
- ↑ The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.
- ↑ 11.0 11.1 Szalai AJ. The biological functions of C-reactive protein. Vascul Pharmacol. 2002 Aug;39(3):105-7. PMID:12616974
