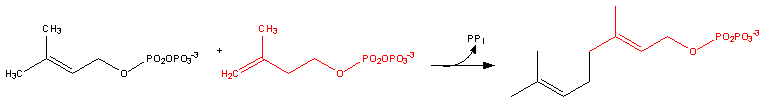Sandbox Reserved 1132
From Proteopedia
| Line 4: | Line 4: | ||
== Family == | == Family == | ||
| - | FPPS is a member of the transferase class and the Trans-Isoprenyl Diphosphate Synthases family (Trans_IPPS). Trans_IPPS family of synthases either synthasases geranyl, farnesyl diphosphates or longer chained products. They produce many different products as steroids, cholesterol, carotenoids, ubiquinone, diterpenes and their precursors. The protein family is widespread among archae, bacteria and eukaryotes and shows a high degree of evolutionary conservation. The “isoprenoid synthase fold”, which each member of this protein family contains, enables them to form linear isoprene chains or isoprenoid diphosphates. Another characteristic element of this family are aspartate-rich, antiparallel alpha helices, which form a large central cavity, containing the catalytic side. Other members of this family are for example Geranylgeranyl pyrophosphate synthase (GGPS1) and Farnesyl pyrophosphate synthase (FDPS). | + | FPPS is a member of the transferase class and the Trans-Isoprenyl Diphosphate Synthases family (Trans_IPPS). Trans_IPPS family of synthases either synthasases geranyl, farnesyl diphosphates or longer chained products. They produce many different products as steroids, cholesterol, carotenoids, ubiquinone, diterpenes and their precursors. The protein family is widespread among archae, bacteria and eukaryotes and shows a high degree of evolutionary conservation. The “isoprenoid synthase fold”, which each member of this protein family contains, enables them to form linear isoprene chains or isoprenoid diphosphates. Another characteristic element of this family are aspartate-rich, antiparallel alpha helices, which form a large central cavity, containing the catalytic side. Other members of this family are for example Geranylgeranyl pyrophosphate synthase (GGPS1) and Farnesyl pyrophosphate synthase (FDPS). <ref name = "rudy" > http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?hslf=1&uid=cd00867&#seqhrch </ref> |
== Structure == | == Structure == | ||
| Line 15: | Line 15: | ||
The side chain is located in the hydrophobic cleft that normally accomodates an isoprenoid lipid, and the phosphonate groups are bound to a cluster of three Mg<sup>2+</sup> ions, which were chelated by two asparate-rich motifs. | The side chain is located in the hydrophobic cleft that normally accomodates an isoprenoid lipid, and the phosphonate groups are bound to a cluster of three Mg<sup>2+</sup> ions, which were chelated by two asparate-rich motifs. | ||
| - | There is a large cavity in the helical bundle that forms a partly hydrophobic ligand-binding site. This cavity is delimited by Phe-113. <ref name = "paper1" </ref> | + | There is a large cavity in the helical bundle that forms a partly hydrophobic ligand-binding site. This cavity is delimited by Phe-113. <ref name = "paper1" > </ref> |
N-BP binding causes a structural rearrangement and a decrease of size in the internal cavity. The movement is mediated by α4 and α8 which participate in the ligand binding. Many connections occur in this movement : residues 249-268 bind over the N-BP and stay in place because of polar interactions : loop residue Lys-257 contacts Asp-243 and a phosphonate oxygen. Asp-247 at the end of α8 forms bidentate hydrogen bonds with main-chain amides of Thr-260 and Asp-261. The main chain from Ile-258 to Thr-260 is separated. The heterocyclic ring structures of RIS and ZOL are surrounded mainly by hydrophobic side chains of residues Phe-99, Leu-100, Thr-167, Lys-200, and Tyr-204, and the nitrogen atom of the ring system is found within hydrogen-bonding. There is a network of salt links involving Lys-57, the terminal carboxylate of Lys-353, and Arg-351. | N-BP binding causes a structural rearrangement and a decrease of size in the internal cavity. The movement is mediated by α4 and α8 which participate in the ligand binding. Many connections occur in this movement : residues 249-268 bind over the N-BP and stay in place because of polar interactions : loop residue Lys-257 contacts Asp-243 and a phosphonate oxygen. Asp-247 at the end of α8 forms bidentate hydrogen bonds with main-chain amides of Thr-260 and Asp-261. The main chain from Ile-258 to Thr-260 is separated. The heterocyclic ring structures of RIS and ZOL are surrounded mainly by hydrophobic side chains of residues Phe-99, Leu-100, Thr-167, Lys-200, and Tyr-204, and the nitrogen atom of the ring system is found within hydrogen-bonding. There is a network of salt links involving Lys-57, the terminal carboxylate of Lys-353, and Arg-351. | ||
| Line 32: | Line 32: | ||
| - | Dimethylallyl diphosphate + isopentenyl diphosphate = diphosphate + geranyl diphosphate< | + | Dimethylallyl diphosphate + isopentenyl diphosphate = diphosphate + geranyl diphosphate <ref name = "image1"> https://en.wikipedia.org/wiki/Farnesyl_pyrophosphate#/media/File:Cholesterol-Synthesis-Reaction8.png </ref> |
| Line 40: | Line 40: | ||
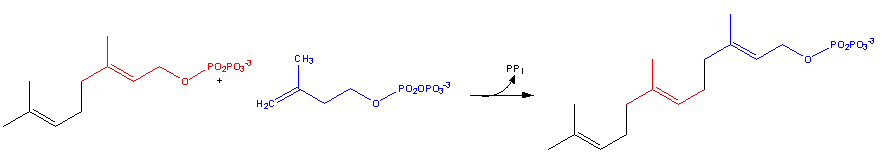
| - | Geranyl diphosphate + isopentenyl diphosphate = diphosphate + (2E,6E)-farnesyl diphosphate< | + | Geranyl diphosphate + isopentenyl diphosphate = diphosphate + (2E,6E)-farnesyl diphosphate <ref name = "image2"> https://en.wikipedia.org/wiki/Farnesyl_pyrophosphate#/media/File:Cholesterol-Synthesis-Reaction9.png </ref> |
====Mevalonate pathway==== | ====Mevalonate pathway==== | ||
Revision as of 13:51, 28 January 2016
Bold text
|
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Human Farnesyl pyrophoasphate synthase (FPPS) is a transferase, which belongs to the Trans-Isoprenyl Diphosphate Synthases family. It is an important enzyme in isoprenoid synthesis, which catalyses the formation of farmesyl diphosphate (FPP). FPP is a precursor for important metabolizes like sterols, dolichols and ubiquinones.
Contents |
Family
FPPS is a member of the transferase class and the Trans-Isoprenyl Diphosphate Synthases family (Trans_IPPS). Trans_IPPS family of synthases either synthasases geranyl, farnesyl diphosphates or longer chained products. They produce many different products as steroids, cholesterol, carotenoids, ubiquinone, diterpenes and their precursors. The protein family is widespread among archae, bacteria and eukaryotes and shows a high degree of evolutionary conservation. The “isoprenoid synthase fold”, which each member of this protein family contains, enables them to form linear isoprene chains or isoprenoid diphosphates. Another characteristic element of this family are aspartate-rich, antiparallel alpha helices, which form a large central cavity, containing the catalytic side. Other members of this family are for example Geranylgeranyl pyrophosphate synthase (GGPS1) and Farnesyl pyrophosphate synthase (FDPS). [1]
Structure
FPPS is a dimer made of two identical subunits, which each contain 13 α-helices and connecting loops. The regions which connect the helices α4-α5 and α8-α9 are extended. Within the handle of helices FPPS has a large central cavity, which acts as a hydrophobic ligand-binding site. The site-chain of Phe-113 limits the cavity on one site. The helices α4 and α8 contain highly conserved and aspartate-rich motives (103DDIUD107 and 243DDYLD247). A distinctive kink in helix α7 turns carbonyl of Lys-200 in the direction of the cavity.[2]
Biological Function and Diseases
Interactions
Interactions with N-BP
The side chain is located in the hydrophobic cleft that normally accomodates an isoprenoid lipid, and the phosphonate groups are bound to a cluster of three Mg2+ ions, which were chelated by two asparate-rich motifs.
There is a large cavity in the helical bundle that forms a partly hydrophobic ligand-binding site. This cavity is delimited by Phe-113. [2]
N-BP binding causes a structural rearrangement and a decrease of size in the internal cavity. The movement is mediated by α4 and α8 which participate in the ligand binding. Many connections occur in this movement : residues 249-268 bind over the N-BP and stay in place because of polar interactions : loop residue Lys-257 contacts Asp-243 and a phosphonate oxygen. Asp-247 at the end of α8 forms bidentate hydrogen bonds with main-chain amides of Thr-260 and Asp-261. The main chain from Ile-258 to Thr-260 is separated. The heterocyclic ring structures of RIS and ZOL are surrounded mainly by hydrophobic side chains of residues Phe-99, Leu-100, Thr-167, Lys-200, and Tyr-204, and the nitrogen atom of the ring system is found within hydrogen-bonding. There is a network of salt links involving Lys-57, the terminal carboxylate of Lys-353, and Arg-351.
IPP inhibits FPPS by bindind to the DMAPP/GPP site, and so N-BP competes with it but the IPP binding site is likely to be the substrate inhibition site, named GPP site. The inhibition compete with GPP, forming an enzyme-inhibitor complex. Furthemore, N-BP binding is magnesium-dependent and occurs in the DMAPP/GPP site.
Interactions with Viperin
Intracellular interaction of viperin with FPPS decreased the activity of the enzyme. Overexpression of FPPS reversed viperin-mediated inhibition of virus production and restored normal membrane fluidity. The viperin-induced inhibition of influenza virus release is mediated by its ability to bind FPPS. [3]
Pathway
FPPS catalyses the reaction of the precursor of the following reactions.
Dimethylallyl diphosphate + isopentenyl diphosphate = diphosphate + geranyl diphosphate [4]
Geranyl diphosphate + isopentenyl diphosphate = diphosphate + (2E,6E)-farnesyl diphosphate [5]
Mevalonate pathway
The mevalonate pathway begins with acetyl-CoA and ends with the production of IPP and DMAPP. It is best known as the target of statins, a class of cholesterol lowering drugs.
Non-mevalonate pathway
The non-mevalonate pathway is an alternative metabolic pathway leading to the formation of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP).
</StructureSection>
References
- ↑ http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?hslf=1&uid=cd00867&#seqhrch
- ↑ 2.0 2.1 Kavanagh KL, Guo K, Dunford JE, Wu X, Knapp S, Ebetino FH, Rogers MJ, Russell RG, Oppermann U. The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs. Proc Natl Acad Sci U S A. 2006 May 16;103(20):7829-34. Epub 2006 May 9. PMID:16684881
- ↑ Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007 Aug 16;2(2):96-105. PMID:18005724 doi:http://dx.doi.org/10.1016/j.chom.2007.06.009
- ↑ https://en.wikipedia.org/wiki/Farnesyl_pyrophosphate#/media/File:Cholesterol-Synthesis-Reaction8.png
- ↑ https://en.wikipedia.org/wiki/Farnesyl_pyrophosphate#/media/File:Cholesterol-Synthesis-Reaction9.png