We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1126
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
=Human cystathionine β-synthase (hCBS)= | =Human cystathionine β-synthase (hCBS)= | ||
| - | <StructureSection load='4L0D' size='500' side='right' caption='Structure of CBS' scene=''> | + | <StructureSection load='4L0D' size='500' side='right' caption='Structure of the dimer form of CBS' scene=''> |
== Introduction == | == Introduction == | ||
| Line 13: | Line 13: | ||
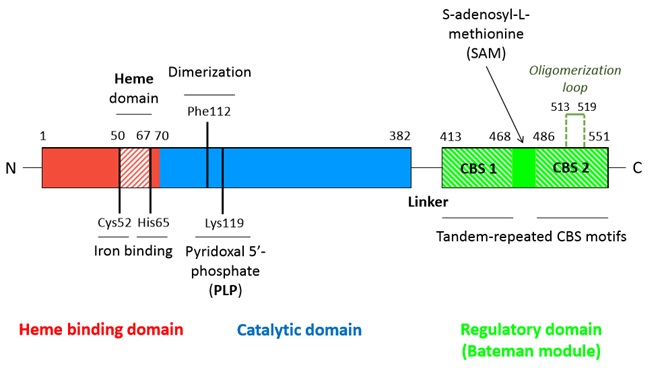
A CBS monomer is natively a 63 kDa protein made of 551 amino-acids and each of them binds two cofactors (the iron heme and the pyridoxal phosphate), as well as two substrates (homocysteine and serine). Hence, the CBS contains (from N-terminal to C-terminal): <br/> | A CBS monomer is natively a 63 kDa protein made of 551 amino-acids and each of them binds two cofactors (the iron heme and the pyridoxal phosphate), as well as two substrates (homocysteine and serine). Hence, the CBS contains (from N-terminal to C-terminal): <br/> | ||
| - | - a heme binding site located in a hydrophobic pocket (residues 50-67) <br/> | + | - a <scene name='71/719867/Scene_4/2'>heme iron</scene> binding site located in a hydrophobic pocket (residues 50-67) <br/> |
| - | - a pyridoxal phosphate (covalently linked to Lysine 119 amino group) situated in a highly conserved central catalytic domain (residues 70-382)<br/> | + | - a pyridoxal phosphate = <scene name='71/719867/Scene_4/1'>PLP</scene> (covalently linked to Lysine 119 amino group) situated in a highly conserved central catalytic domain (residues 70-382)<br/> |
| - | - a C-terminal regulatory domain called Bateman module composed of two CBS domains (CBS1 and CBS2) | + | - a <scene name='71/719867/Scene_2/1'>C-terminal regulatory domain</scene> called Bateman module composed of two CBS domains (CBS1 and CBS2) |
[[Image:Linear_structure_CBS_monomer.jpg]] | [[Image:Linear_structure_CBS_monomer.jpg]] | ||
| Line 21: | Line 21: | ||
==== Cofactor ==== | ==== Cofactor ==== | ||
| - | *Heme iron: | + | *<scene name='71/719867/Scene_4/2'>Heme iron</scene>: |
The heme is one of the two cofactors of hCBS. | The heme is one of the two cofactors of hCBS. | ||
It is bound in an hydrophobic pocket composed of the residues 50-67. The iron atom is hexacoordinated with the sulfhydryl group of Cys52 and the Nε2 atom of His65 (axial coordination) and with the four nitrogen atoms of the heme. | It is bound in an hydrophobic pocket composed of the residues 50-67. The iron atom is hexacoordinated with the sulfhydryl group of Cys52 and the Nε2 atom of His65 (axial coordination) and with the four nitrogen atoms of the heme. | ||
| Line 27: | Line 27: | ||
It is supposed to act as a redox sensor or as a way to facilitate a correct folding. | It is supposed to act as a redox sensor or as a way to facilitate a correct folding. | ||
| - | *Pyridoxal phosphate: | + | *<scene name='71/719867/Scene_4/1'>Pyridoxal phosphate (PLP)</scene>: |
| Line 36: | Line 36: | ||
*Dimer formation: two monomers shape a dimer through both hydrophobic and polar interactions within the catalytic core. Hydrophobic interactions particularly involve two Phe112 (one from each monomer) which interact with each other. There are no interactions between Bateman modules in a single dimer concerning hCBS. | *Dimer formation: two monomers shape a dimer through both hydrophobic and polar interactions within the catalytic core. Hydrophobic interactions particularly involve two Phe112 (one from each monomer) which interact with each other. There are no interactions between Bateman modules in a single dimer concerning hCBS. | ||
| - | * Tetramer formation involves the Bateman modules as well as the catalytic core of each dimer. Each oligomerization loop (loop 513-519) of a monomer of one dimer interacts with the catalytic core of a monomer of the other dimer. Those loops interact within a crevace (shaped by α-helix 5-6-12-15-16 and β-strands 5-6) of the catalytic core. The tetramer is an inactive form of the enzyme. | + | * Tetramer formation involves the Bateman modules as well as the catalytic core of each dimer. Each <scene name='71/719867/Scene_3/1'>oligomerization loop</scene> (loop 513-519) of a monomer of one dimer interacts with the catalytic core of a monomer of the other dimer. Those loops interact within a crevace (shaped by α-helix 5-6-12-15-16 and β-strands 5-6) of the catalytic core. The tetramer is an inactive form of the enzyme. |
Revision as of 19:02, 29 January 2016
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Human cystathionine β-synthase (hCBS)
| |||||||||||