We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1136
From Proteopedia
(Difference between revisions)
| Line 32: | Line 32: | ||
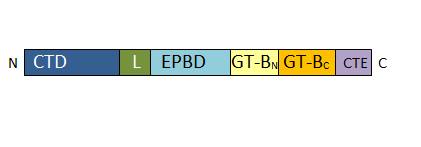
• 157-276: EPBD: ENOD40 peptide-binding domain. This domain has a role in the regulation of the enzyme. It is able to bind a potassium ion. | • 157-276: EPBD: ENOD40 peptide-binding domain. This domain has a role in the regulation of the enzyme. It is able to bind a potassium ion. | ||
| - | • 277-776 : GT-B glycosyltransferase domain. It contains the | + | • 277-776 : GT-B glycosyltransferase domain. It contains the catalytic site and presents a characteristic Rossman-folding |
<ref>Glycosyltransferases: structures, functions, and mechanisms. Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu Rev Biochem. 2008 | <ref>Glycosyltransferases: structures, functions, and mechanisms. Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu Rev Biochem. 2008 | ||
</ref> | </ref> | ||
| Line 38: | Line 38: | ||
• 776-808 : C-terminal extension. The length of this domain is variable depending of the SUS isoform. | • 776-808 : C-terminal extension. The length of this domain is variable depending of the SUS isoform. | ||
| - | [[Image: | + | [[Image:Monomer structure.jpg]] |
| - | The SUS1 tetramer is flat, with two types of subunit interfaces, the A:B and A:D interfaces. The A:D interface is an interaction between the C-terminal extension and the linker, whereas the A:B interface is created by the interaction of | + | The SUS1 tetramer is flat, with two types of subunit interfaces, the A:B and A:D interfaces. The A:D interface is an interaction between the C-terminal extension and the linker, whereas the A:B interface is created by the interaction of adjacent EPBD domains. |
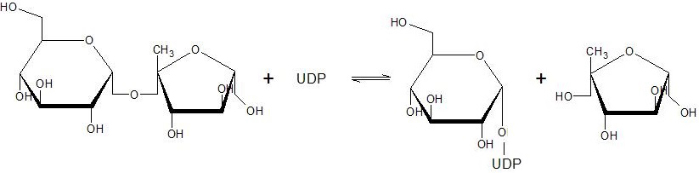
The active site of SUS1 is able to bind both fructose and UDP-glucose. UDP-glucose is mainly bound by the | The active site of SUS1 is able to bind both fructose and UDP-glucose. UDP-glucose is mainly bound by the | ||
Revision as of 13:28, 30 January 2016
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Structure and Functional aspects of Sucrose Synthase from Arabidopsis thaliana
Sucrose Synthase 1 (EC:2.4.1.13), also known as the Sucrose-UDP glucolsyltransferase 1, is a reversible enzyme allowing the synthesis or the degradation of Sucrose in Arabidopsis thaliana. It is a 360 kDa tetramer and belongs to the Glycosyltransferase subfamily 4 (GT4).
| |||||||||||
References
• UniProt entry: P49040
• Brenda entry : 2.4.1.13
- ↑ Salerno GL, Curatti L Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci. 2003 Feb
- ↑ Baroja-Fernández, E., Muñoz, F.J., Saikusa, T., Rodríguez-López, M., Akazawa, T. and Pozueta-Romero, J. Sucrose synthase catalyzes the de novo production of ADPglucose linked to starch biosynthesis in heterotrophic tissues of plants. Plant Cell Physiol.
- ↑ Sucrose synthase oligomerization and F-actin association are regulated by sucrose concentration and phosphorylation. Duncan KA, Huber SC. Plant Cell Physiol. 2007 Nov; 48(11):1612-23.
- ↑ Salerno GL, Curatti L. Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci. 2003 Feb
- ↑ Determination of structural requirements and probable regulatory effectors for membrane association of maize sucrose synthase 1. Hardin SC, Duncan KA, Huber SC. Plant Physiol. 2006
- ↑ Phosphorylation of sucrose synthase at serine 170: occurrence and possible role as a signal for proteolysis. Hardin SC, Tang GQ, Scholz A, Holtgraewe D, Winter H, Huber SC. Plant J. 2003
- ↑ Glycosyltransferases: structures, functions, and mechanisms. Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu Rev Biochem. 2008