This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1131
From Proteopedia
| Line 32: | Line 32: | ||
===Catalyic mechanism=== | ===Catalyic mechanism=== | ||
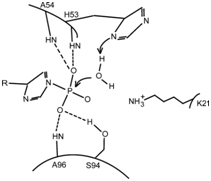
The P(i) substrate binds to the active site thanks to the formation of four H-bonds. The imidazole ring of His(53) acts as a base to activate a water molecule, which will in turn do a nucleophilic attack on the substrate. The amine group of the residue Lys(21) can help stabilize the transition state. | The P(i) substrate binds to the active site thanks to the formation of four H-bonds. The imidazole ring of His(53) acts as a base to activate a water molecule, which will in turn do a nucleophilic attack on the substrate. The amine group of the residue Lys(21) can help stabilize the transition state. | ||
| + | [[Image:PHPT1 catalytic mechanism.jpg|300px|left|thumb| PHPT1 catalytic mechanism]] | ||
= Implication in biological fonctions and pathways = | = Implication in biological fonctions and pathways = | ||
Revision as of 16:47, 30 January 2016
|
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
(Human) Phosphohistidine phosphatase 1 belongs to the Janus family, and has 2 isoforms produced by alternative splicing, and 6 transcripts. It is encoded by the PHPT1 gene, located on the 9th chromosome.
Contents |
Structure
|
Generality
Phosphohistidine phosphatase 1 is a 14kDa homotrimeric protein, which monomers contain all 125 amino acids. Furthermore, one monomer contains 4 α helices, 6 β strands and 2 turns.
Domains
PHPT1 contains acetylation site and a N-acetylalanine site. There is also an Janus/Ocnus family region, characteristic.
It has one substrate binding site, and one proton acceptor active site.
Function
It is an hydrolase. This characteristic structure allows it to have many activities : phosphoprotein and phosphohistidine phosphatase, calcium channel inhibition, ion channel binding. It is located in the cytosol and in extracellular exosome.
Active site
The active site is located between the beginning of helix α1 and loop L5. The catalytic residue is His(53), and the anchor sites of P(i) are the amine groups of His(53), Ala(54) and Ala(96) and the Hydroxy group of Ser(94), which form hydrogen bonds with it.
Ligands and binding
There are 6 SO4 lingands, called SO4 201 to 206. 2 lingand fix to each monomer (2ai6), inducing a conformational change of all the monomers (2hw4). Then, the lingands bind to each other, forming a trimer (2NMM). O4 S is a 96 Da molecule, with two negative charges. The four oxygens allow good fixation on the monomers, with hydrogen bounds.
Catalyic mechanism
The P(i) substrate binds to the active site thanks to the formation of four H-bonds. The imidazole ring of His(53) acts as a base to activate a water molecule, which will in turn do a nucleophilic attack on the substrate. The amine group of the residue Lys(21) can help stabilize the transition state.
Implication in biological fonctions and pathways
Diseases and medical applications
Structural highlights
</StructureSection>