We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1126
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
== Introduction == | == Introduction == | ||
| - | The human cystathionine β-synthase (hCBS) is natively a homotetrameric enzyme (EC 4.2.1.22) catalyzing the following reaction (which is part of the | + | The human cystathionine β-synthase (hCBS) is natively a homotetrameric enzyme (EC 4.2.1.22) catalyzing the following reaction (which is part of the cysteine biosynthesis pathway) using homocysteine and serine <ref name="structure of">PMID: 11483494</ref> : |
IMAGE <br/> | IMAGE <br/> | ||
It is encoded by the CBS gene located on chromosome 21 (at position 22.3). | It is encoded by the CBS gene located on chromosome 21 (at position 22.3). | ||
| Line 12: | Line 12: | ||
== Structure == | == Structure == | ||
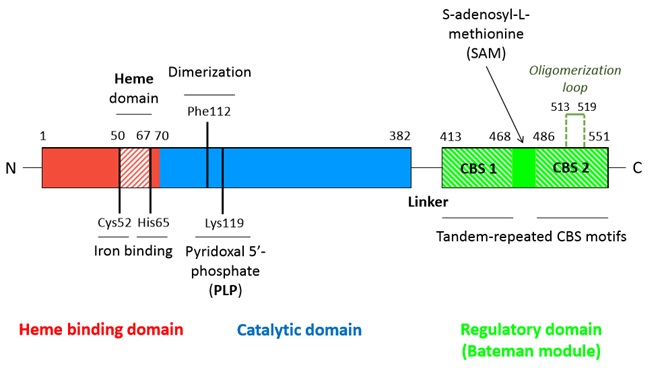
| - | A CBS monomer is a | + | A CBS monomer is a 551 amino-acids protein of 63 kDa.Each monomer binds two cofactors (the iron heme and the pyridoxal phosphate), as well as two substrates (homocysteine and serine) <ref name="structure of"/>. <br/> Hence, a CBS monomer contains (from N-terminal to C-terminal): <br/> |
- a <scene name='71/719867/Scene_4/2'>heme iron</scene> binding site located in a hydrophobic pocket (residues 50-67), <br/> | - a <scene name='71/719867/Scene_4/2'>heme iron</scene> binding site located in a hydrophobic pocket (residues 50-67), <br/> | ||
- a pyridoxal phosphate = <scene name='71/719867/Scene_4/1'>PLP</scene> (covalently linked to Lysine 119 amino group) situated in a <scene name='71/719867/Scene_1/1'>highly conserved catalytic domain</scene> (residues 70-382), <br/> | - a pyridoxal phosphate = <scene name='71/719867/Scene_4/1'>PLP</scene> (covalently linked to Lysine 119 amino group) situated in a <scene name='71/719867/Scene_1/1'>highly conserved catalytic domain</scene> (residues 70-382), <br/> | ||
| Line 23: | Line 23: | ||
*<scene name='71/719867/Scene_4/2'>Heme iron</scene> | *<scene name='71/719867/Scene_4/2'>Heme iron</scene> | ||
The heme is one of the two cofactors of hCBS. | The heme is one of the two cofactors of hCBS. | ||
| - | It | + | It binds an hydrophobic pocket composed of the residues 50-67. The iron atom is hexacoordinated with the sulfhydryl group of Cys52 and the Nε2 atom of His65 (axial coordination) and with the four nitrogen atoms of the heme. |
Although the heme is essential for activity in human CBS, its role still remains an enigma. | Although the heme is essential for activity in human CBS, its role still remains an enigma. | ||
It is supposed to act as a redox sensor or as a way to facilitate a correct folding. | It is supposed to act as a redox sensor or as a way to facilitate a correct folding. | ||
| Line 32: | Line 32: | ||
==== Quaternary structure ==== | ==== Quaternary structure ==== | ||
| - | The hCBS is natively a homotetrameric enzyme. It | + | The hCBS is natively a homotetrameric enzyme. It has been suggested that two monomers form a dimer, and then two dimers form a tetramer. |
* '''Dimer formation''' | * '''Dimer formation''' | ||
| - | Two monomers | + | Two monomers bind together to form a dimer through both hydrophobic and polar interactions within the catalytic core of each monomer. Hydrophobic interactions particularly involve two Phe112 (one from each monomer) which interact with each other. For hCBS there are no interactions between Bateman modules in a single dimer. |
* '''Tetramer formation''' | * '''Tetramer formation''' | ||
| - | It involves the Bateman modules as well as the catalytic cores of each dimer. Each <scene name='71/719867/Scene_3/1'>oligomerization loop</scene> (loop 513-519) of a monomer of one dimer interacts with the catalytic core of a monomer of the other dimer. Those loops interact within a | + | It involves the Bateman modules as well as the catalytic cores of each dimer. Each <scene name='71/719867/Scene_3/1'>oligomerization loop</scene> (loop 513-519) of a monomer of one dimer interacts with the catalytic core of a monomer of the other dimer. Those loops interact within a cavity (shaped by α-helix 5-6-12-15-16 and β-strands 5-6) of the catalytic core. The tetramer is an inactive form of the enzyme. |
| Line 56: | Line 56: | ||
- '''hydrophobic interactions''' between residues I537, L540, A544 of the CBS1 domain of the <scene name='71/719867/Scene_2/1'>Bateman module</scene> of one monomer with residues I166, V189, V206, L210, I214 of the <scene name='71/719867/Scene_1/1'>catalytic core</scene> of the other monomer, <br/> | - '''hydrophobic interactions''' between residues I537, L540, A544 of the CBS1 domain of the <scene name='71/719867/Scene_2/1'>Bateman module</scene> of one monomer with residues I166, V189, V206, L210, I214 of the <scene name='71/719867/Scene_1/1'>catalytic core</scene> of the other monomer, <br/> | ||
- '''hydrogen bounds''' between residues T460, N463, S466, Y484 of the CBS2 domain of the <scene name='71/719867/Scene_2/1'>Bateman module</scene> of one monomer with residues E201, N194, R196, D198 of the loop 191-202 of the <scene name='71/719867/Scene_1/1'>catalytic core</scene> of the other monomer. <br/><br/> | - '''hydrogen bounds''' between residues T460, N463, S466, Y484 of the CBS2 domain of the <scene name='71/719867/Scene_2/1'>Bateman module</scene> of one monomer with residues E201, N194, R196, D198 of the loop 191-202 of the <scene name='71/719867/Scene_1/1'>catalytic core</scene> of the other monomer. <br/><br/> | ||
| - | These hydrophobic interactions combined with the hydrogen | + | These hydrophobic interactions, combined with the hydrogen bound network '''anchor the Bateman module of one monomer to the entrance of the catalytic core of the other monomer''' (close conformation), thus making it impossible for any substrate to get access it. |
==== Activation by S-adenosyl-L-methionine (SAM) ==== | ==== Activation by S-adenosyl-L-methionine (SAM) ==== | ||
| - | *SAM is the '''allosteric activator''' of the CBS. It binds in a region located between the CBS1 and CBS2 domains of the Bateman module which is solvent-exposed and has less hefty hydrophobic residues. In addition, this region | + | *SAM is the '''allosteric activator''' of the CBS. It binds in a region located between the CBS1 and CBS2 domains of the Bateman module which is solvent-exposed and has less hefty hydrophobic residues. In addition, this region forms a hydrophobic cage able to host the adenine ring. Moreover threonine (T535) and aspartate (D538) help to stabilize the ribose through hydrogen bounds and polar interactions. |
*Binding of SAM to the Bateman module destabilizes the interactions which sustain the tetramer structure and thus triggers the '''dissociation of the tetrameric structure''' into two dimers. | *Binding of SAM to the Bateman module destabilizes the interactions which sustain the tetramer structure and thus triggers the '''dissociation of the tetrameric structure''' into two dimers. | ||
| - | *SAM fixation on the C-terminal regulatory domain | + | *SAM fixation on the C-terminal regulatory domain triggers the small rotation (or at least displacement) of the CBS1 and CBS2 of the Bateman module, thus distabilizing its interactions (hydrophobic interactions and hydrogen bounds) with the catalytic core of the other monomer. As a result, '''the Bateman module moves away from the catalytic core''' (this movement is facilitated as the linker region is long enough and made of flexible residues). <scene name='71/719867/Scene_5/1'>Loops 145-148, 171-174 and 191-202</scene>, previously involved in maintaining the close conformation through their interaction with the Bateman module, then relax and therefore '''allow accessibility to the catalytic site''' (open conformation). |
*SAM release leads to the return to the native inactive close conformation. | *SAM release leads to the return to the native inactive close conformation. | ||
| Line 72: | Line 72: | ||
A thight control of cystathionine β-synthase level and activity is crucial for optimal cognitive function. | A thight control of cystathionine β-synthase level and activity is crucial for optimal cognitive function. | ||
| - | Due to the fact that the CBS gene is located on chromosome 21, an '''overexpression of CBS''' is observed in | + | Due to the fact that the CBS gene is located on chromosome 21 (21q22.3), an '''overexpression of CBS''' is observed in patients with '''Down Syndrome''', or trisomy 21. <br/> Hence, Down Syndrome is characterized by high plasma levels of cystathionine and cysteine and this disorder is typically associated with mental retardation. |
| - | On the contrary, a '''reduced activity of CBS''' leads to '''homocystinuria'''. This disorder is inherited in an autosomal recessive pattern and is caused by mutations in the gene | + | On the contrary, a '''reduced activity of CBS''' leads to '''homocystinuria'''. This disorder is inherited in an autosomal recessive pattern and is caused by loss-of-function mutations in the hCBS gene. Those mutations interfere with the activation of CBS. <br/> Thus, Homocystinuria is characterized by high plasma levels of the toxic amino acid homocysteine and infants who develop this disease may have difficulties to grow and gain weight accompanied by mental retardation. |
Revision as of 17:10, 30 January 2016
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Human cystathionine β-synthase (hCBS)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Ereno-Orbea J, Majtan T, Oyenarte I, Kraus JP, Martinez-Cruz LA. Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration. Proc Natl Acad Sci U S A. 2013 Sep 16. PMID:24043838 doi:10.1073/pnas.1313683110