Sandbox Reserved 1126
From Proteopedia
(Difference between revisions)
| Line 64: | Line 64: | ||
- '''hydrophobic interactions''' between residues I537, L540, A544 of the CBS1 domain of the <scene name='71/719867/Scene_2/1'>Bateman module</scene> of one monomer with residues I166, V189, V206, L210, I214 of the <scene name='71/719867/Scene_1/1'>catalytic core</scene> of the other monomer, <br/> | - '''hydrophobic interactions''' between residues I537, L540, A544 of the CBS1 domain of the <scene name='71/719867/Scene_2/1'>Bateman module</scene> of one monomer with residues I166, V189, V206, L210, I214 of the <scene name='71/719867/Scene_1/1'>catalytic core</scene> of the other monomer, <br/> | ||
- '''hydrogen bounds''' between residues T460, N463, S466, Y484 of the CBS2 domain of the <scene name='71/719867/Scene_2/1'>Bateman module</scene> of one monomer with residues E201, N194, R196, D198 of the <scene name='71/719867/Scene_5/2'>loop 191-202</scene> of the <scene name='71/719867/Scene_1/1'>catalytic core</scene> of the other monomer. <br/><br/> | - '''hydrogen bounds''' between residues T460, N463, S466, Y484 of the CBS2 domain of the <scene name='71/719867/Scene_2/1'>Bateman module</scene> of one monomer with residues E201, N194, R196, D198 of the <scene name='71/719867/Scene_5/2'>loop 191-202</scene> of the <scene name='71/719867/Scene_1/1'>catalytic core</scene> of the other monomer. <br/><br/> | ||
| - | These hydrophobic interactions, combined with the hydrogen bound network '''anchor the Bateman module of one monomer to the entrance of the catalytic core of the other monomer''' (close conformation), thus making it impossible for any substrate to get access it. | + | These hydrophobic interactions, combined with the hydrogen bound network '''anchor the Bateman module of one monomer to the entrance of the catalytic core of the other monomer''' (close conformation), thus making it impossible for any substrate to get access to it. |
Revision as of 21:32, 30 January 2016
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
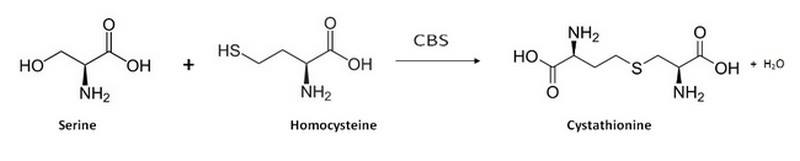
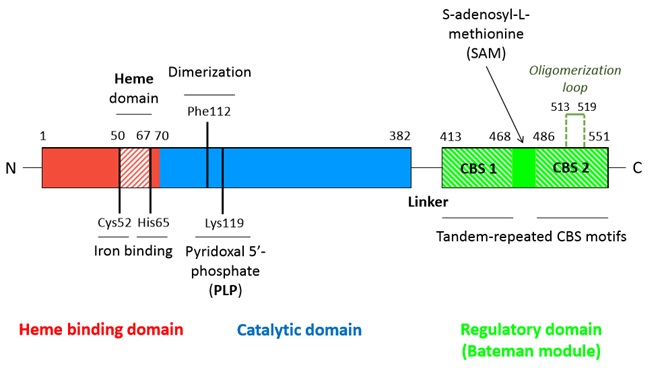
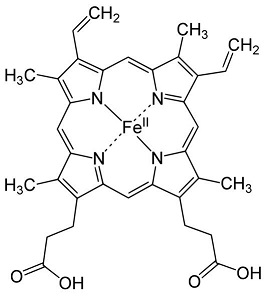
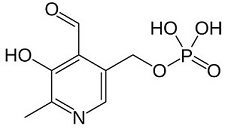
Human cystathionine β-synthase (hCBS)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Regnier V, Billard JM, Gupta S, Potier B, Woerner S, Paly E, Ledru A, David S, Luilier S, Bizot JC, Vacano G, Kraus JP, Patterson D, Kruger WD, Delabar JM, London J. Brain phenotype of transgenic mice overexpressing cystathionine beta-synthase. PLoS One. 2012;7(1):e29056. doi: 10.1371/journal.pone.0029056. Epub 2012 Jan 12. PMID:22253703 doi:http://dx.doi.org/10.1371/journal.pone.0029056
- ↑ McCorvie TJ, Kopec J, Hyung SJ, Fitzpatrick F, Feng X, Termine D, Strain-Damerell C, Vollmar M, Fleming J, Janz JM, Bulawa C, Yue WW. Inter-Domain Communication Of Human Cystathionine Beta Synthase: Structural Basis Of S-Adenosyl-L-Methionine Activation. J Biol Chem. 2014 Oct 21. pii: jbc.M114.610782. PMID:25336647 doi:http://dx.doi.org/10.1074/jbc.M114.610782
- ↑ 4.0 4.1 4.2 Ereno-Orbea J, Majtan T, Oyenarte I, Kraus JP, Martinez-Cruz LA. Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration. Proc Natl Acad Sci U S A. 2013 Sep 16. PMID:24043838 doi:10.1073/pnas.1313683110
- ↑ Miles EW, Kraus JP. Cystathionine beta-synthase: structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem. 2004 Jul 16;279(29):29871-4. Epub 2004 Apr 15. PMID:15087459 doi:http://dx.doi.org/10.1074/jbc.R400005200
- ↑ http://www.ebi.ac.uk/interpro/entry/IPR004625
- ↑ https://pubchem.ncbi.nlm.nih.gov/compound/pyridoxal_phosphate#section=Top
- ↑ 8.0 8.1 Ereno-Orbea J, Majtan T, Oyenarte I, Kraus JP, Martinez-Cruz LA. Structural insight into the molecular mechanism of allosteric activation of human cystathionine beta-synthase by S-adenosylmethionine. Proc Natl Acad Sci U S A. 2014 Sep 16;111(37):E3845-52. doi:, 10.1073/pnas.1414545111. Epub 2014 Sep 2. PMID:25197074 doi:http://dx.doi.org/10.1073/pnas.1414545111