We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1127
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Function == | == Function == | ||
| - | PDE5 is 5th isoform of the 12 known phosphodiesterases. This enzyme family is responsible for cyclic AMP and cyclic GMP(cGMP) hydrolysis into AMP and GMP. :PE5 specifically catalyzes the hydrolysis of cGMP into 5'GMP. cGMP is an usual second messager in cell signalling pathways, like NO synthesis. By degradating such nucleotide,this phosphodiesterase of class I regulates nucletotic concentration of the cell, through an allosteric regulation system. | + | PDE5 is 5th isoform of the 12 known phosphodiesterases. This enzyme family is responsible for cyclic AMP and cyclic GMP(cGMP) hydrolysis into AMP and GMP. :PE5 specifically catalyzes the hydrolysis of cGMP into 5'GMP. cGMP is an usual second messager in cell signalling pathways, like NO synthesis. By degradating such nucleotide, this phosphodiesterase of class I regulates nucletotic concentration of the cell, through an allosteric regulation system. |
PDE5 function affects the smooth muscles, blood vessels or penis, uterus and intestines. | PDE5 function affects the smooth muscles, blood vessels or penis, uterus and intestines. | ||
| Line 36: | Line 36: | ||
cGMP is hydrolyzed in GMP. The catalytic chemical reaction deals with breaking a phosphodiester bound in cGMP between the 3'O of the guanoside and the phosphate of cGMP: cGMP + H20 => GMP. | cGMP is hydrolyzed in GMP. The catalytic chemical reaction deals with breaking a phosphodiester bound in cGMP between the 3'O of the guanoside and the phosphate of cGMP: cGMP + H20 => GMP. | ||
| - | The catalytic domain is able to bind ligands thanks to an <scene name='71/719868/Hydro/1'>hydrophobic pocket</scene> (<span style="color:magenta;background-color:black;font-weight:bold;">hydrophylic domain</span>. Moreover with this <scene name='71/719868/Conserve/1'>animation scene</scene> it appears the cGMP binding site in the catalytic domain is <span style="color:darkred;background-color:black;font-weight:bold;">more conserved</span>than the the rest of the domain that could be <span style="color:lightseagreen;background-color:black;font-weight:bold;">variable</span>. <span style="color:yellow;background-color:black;font-weight:bold;">Sildenafil is in yellow</span> | + | The catalytic domain is able to bind ligands thanks to an <scene name='71/719868/Hydro/1'>hydrophobic pocket</scene> (<span style="color:magenta;background-color:black;font-weight:bold;">hydrophylic domain</span>. Moreover with this <scene name='71/719868/Conserve/1'>animation scene</scene> it appears the cGMP binding site in the catalytic domain is <span style="color:darkred;background-color:black;font-weight:bold;">more conserved</span> than the the rest of the domain that could be <span style="color:lightseagreen;background-color:black;font-weight:bold;">variable</span>. <span style="color:yellow;background-color:black;font-weight:bold;">Sildenafil is in yellow</span> |
Some residues could be affected by post traductional modifications : | Some residues could be affected by post traductional modifications : | ||
| Line 42: | Line 42: | ||
acylation : K364. | acylation : K364. | ||
ubiquitinylation : K714. | ubiquitinylation : K714. | ||
| - | The <span style="color:fuchsia;background-color:black;font-weight:bold;">ubiquitinylation)</span> of <scene name='71/719868/K714/1'>K714</scene> and the <span style="color:gold;background-color:black;font-weight:bold;">phosphorylation</span> of <scene name='71/719868/S869/1'>S869</scene> | + | The <span style="color:fuchsia;background-color:black;font-weight:bold;">ubiquitinylation)</span> of <scene name='71/719868/K714/1'>K714</scene> and the <span style="color:gold;background-color:black;font-weight:bold;">phosphorylation</span> of <scene name='71/719868/S869/1'>S869</scene> are present in the catalytic site and the phosphorylation of S819 is involved in cGMP binding. |
== Inhibitors and medical application == | == Inhibitors and medical application == | ||
| Line 55: | Line 55: | ||
==Allosteric regulation== | ==Allosteric regulation== | ||
===Positive regulation=== | ===Positive regulation=== | ||
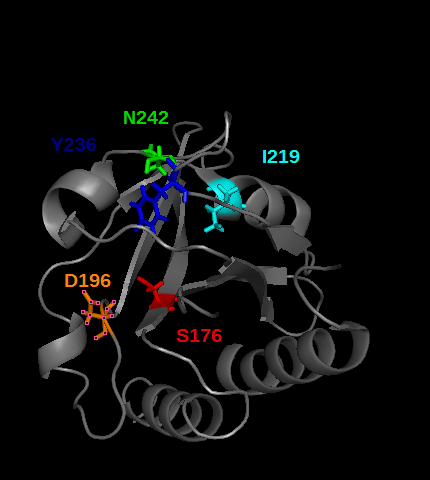
| - | PDE5 contains several cGMP binding sites that do not have the same effect on its activity. These allosteric domains are GAF A and GAF B play a role in PDE5 activity. Especially GAF A(see the picture below) binds cGMP, by establishing | + | PDE5 contains several cGMP binding sites that do not have the same effect on its activity. These allosteric domains are GAF A and GAF B play a role in PDE5 activity. Especially GAF A (see the picture below) binds cGMP, by establishing hydrogen bonds between cGMP and the mainly polar following residues<ref>DOI:10.1074/jbc.M801577200</ref> : Tyr236, Asp242, Ile219, Ser176, Asp296 colored in the picture below (obtained from the modified 2FK3 PDB file<ref>http://www.rcsb.org/pdb/explore/explore.do?structureId=2K31</ref>by using pymol) |
| Line 61: | Line 61: | ||
| - | cGMP binding to GAF A of the PDE5 may trigger an allosteric modification that lead to the seperation of the dimeric catalytic site of the enzyme, so that it frees cGMP access to the catalytic site. This model was proposed considering the homology between | + | cGMP binding to GAF A of the PDE5 may trigger an allosteric modification that lead to the seperation of the dimeric catalytic site of the enzyme, so that it frees cGMP access to the catalytic site. This model was proposed considering the homology between PDE5 and PDE2<ref>DOI:10.1111/j.1476-5381.2010.00977.x</ref> |
This cGMP binding to allosteric sites increases the enzyme affinity to cGMP. | This cGMP binding to allosteric sites increases the enzyme affinity to cGMP. | ||
| - | Plus,the phosphorylation the Ser92 by the PKG kinase<ref>DOI:10.1093/emboj/cdg051</ref> leads to increase the catalytic activity of the enzyme. | + | Plus, the phosphorylation the Ser92 by the PKG kinase<ref>DOI:10.1093/emboj/cdg051</ref> leads to increase the catalytic activity of the enzyme. |
===Negative regulation=== | ===Negative regulation=== | ||
Revision as of 08:46, 31 January 2016
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Human PDE5
Contributors
DJAGO Fabiola, AL BADAWY Kays, CHOI Ji-Hyung
| |||||||||||
References
- ↑ http://www.phosphosite.org/proteinAction?id=1026&showAllSites=true
- ↑ http://www.rcsb.org/pdb/explore/explore.do?pdbId=1TBF
- ↑ http://www.uniprot.org/uniprot/O76074#structure
- ↑ http://www.ebi.ac.uk/interpro/sequencesearch/iprscan5-S20160124-100328-0067-2702087-es
- ↑ Francis SH, Blount MA, Corbin JD. Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol Rev. 2011 Apr;91(2):651-90. doi: 10.1152/physrev.00030.2010. PMID:21527734 doi:http://dx.doi.org/10.1152/physrev.00030.2010
- ↑ Zoraghi R, Corbin JD, Francis SH. Phosphodiesterase-5 Gln817 is critical for cGMP, vardenafil, or sildenafil affinity: its orientation impacts cGMP but not cAMP affinity. J Biol Chem. 2006 Mar 3;281(9):5553-8. Epub 2006 Jan 5. PMID:16407275 doi:http://dx.doi.org/10.1074/jbc.M510372200
- ↑ Heikaus CC, Stout JR, Sekharan MR, Eakin CM, Rajagopal P, Brzovic PS, Beavo JA, Klevit RE. Solution structure of the cGMP binding GAF domain from phosphodiesterase 5: insights into nucleotide specificity, dimerization, and cGMP-dependent conformational change. J Biol Chem. 2008 Aug 15;283(33):22749-59. Epub 2008 Jun 4. PMID:18534985 doi:10.1074/jbc.M801577200
- ↑ http://www.rcsb.org/pdb/explore/explore.do?structureId=2K31
- ↑ Jager R, Schwede F, Genieser HG, Koesling D, Russwurm M. Activation of PDE2 and PDE5 by specific GAF ligands: delayed activation of PDE5. Br J Pharmacol. 2010 Dec;161(7):1645-60. doi: 10.1111/j.1476-5381.2010.00977.x. PMID:20698857 doi:http://dx.doi.org/10.1111/j.1476-5381.2010.00977.x
- ↑ Rybalkin SD, Rybalkina IG, Shimizu-Albergine M, Tang XB, Beavo JA. PDE5 is converted to an activated state upon cGMP binding to the GAF A domain. EMBO J. 2003 Feb 3;22(3):469-78. PMID:12554648 doi:http://dx.doi.org/10.1093/emboj/cdg051
- ↑ Gopal VK, Francis SH, Corbin JD. Allosteric sites of phosphodiesterase-5 (PDE5). A potential role in negative feedback regulation of cGMP signaling in corpus cavernosum. Eur J Biochem. 2001 Jun;268(11):3304-12. PMID:11389733
- ↑ Rybalkin SD, Rybalkina IG, Feil R, Hofmann F, Beavo JA. Regulation of cGMP-specific phosphodiesterase (PDE5) phosphorylation in smooth muscle cells. J Biol Chem. 2002 Feb 1;277(5):3310-7. Epub 2001 Nov 26. PMID:11723116 doi:10.1074/jbc.M106562200